NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
NCBI Discovery Workshops May 15-16 at NLM: Seats still available
NCBI will present a two-day workshop May 15 and 16, 2012, on the NIH campus in Bethesda, Maryland. The course is free and is open to anyone interested in NCBI resources. The four workshops are Sequences, Genomes, and Maps; Proteins, Domains and Structures; NCBI BLAST Services; and Human Variation and Disease Genes. These workshops provide hands-on experience exploring practical examples using tools and databases on the NCBI website. The Discovery Workshops page has more details and a link to register for the course.
Assembly: a Companion to the Genome Database
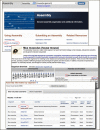
The new NCBI Assembly database provides statistics, update history and links to sequences for eukaryotic genome assemblies including assemblies for previous genome builds. Assemblies of interest can be found either by text searches on the main assembly page or through the assembly browser that provides easy access by organism. Assemblies are also linked through the Genome database main page or from a Genome record for a eukaryotic species as shown in Figure 1. Each assembly is assigned an accession and a version that unambiguously identifies the sequences in a particular version of an assembly. The database contains the placement of each scaffold in the assembly along with the name and sequence accession and version for each chromosome and scaffold. The database also organizes and provides assembly descriptive items such as assembly names and synonyms, as well as statistical reports including scaffold counts and weighted scaffold and contig length medians (N50). Figure 2 shows the page for the latest mouse genome assembly (GRCm38). This page provides access to the primary assembly and alternate loci sequences and statistics. The Assembly Help documentation provides more detailed information on using the Assembly Database.
New Videos on NCBI’s YouTube Channel
Eleven new tutorial videos have been added to the NCBI YouTube channels in the past few months. To make topics of interest easier to find, the Tutorials playlist now provides special playlists for certain resources. The channel now features separate tutorial playlists for Genome Workbench (7 videos), Sequence Viewer (4 videos), My NCBI (4 videos), Genetic Testing Registry (2 videos) and General (22 videos).
Five of the recent videos are in the General playlist and include tutorials on using the new Advanced Search Builder in PubMed (video, advanced search page), an overview of RefSeqGene reference standard records for selected human genes (video, resource page), an introduction to the E-utilities, the programming interface to the Entrez system (video, E-Utilities Help Manual), a demonstration of the highlight sequence features tool in sequence databases (video, NCBI News), and a video on how to use Genome Remapping Tool (video, tool page) that can map coordinates of genes and other markers from one genome build to another.
Three of the new videos are about My NCBI, the service that allows registered users to customize their experience and to save and share results, searches, and preferences through their accounts. New titles in the My NCBI playlist are My Bibliography, Save Searches and Set E-mail Alerts, and Save Search Results in Collections.
One new video demonstrating how load a genome into Genome Workbench was recently added to the playlist for Genome Workbench, NCBI’s standalone sequence analysis and annotation platform.
Most recently a new playlist was created for two tutorials (GTR: Homepage and Basic Search Functions and GTR: Locate a Test in Under Three Minutes) featuring the newly launched Genetic Testing Registry, a repository of information about available genetic tests. Additional information about the GTR is provided in the following section of this newsletter.
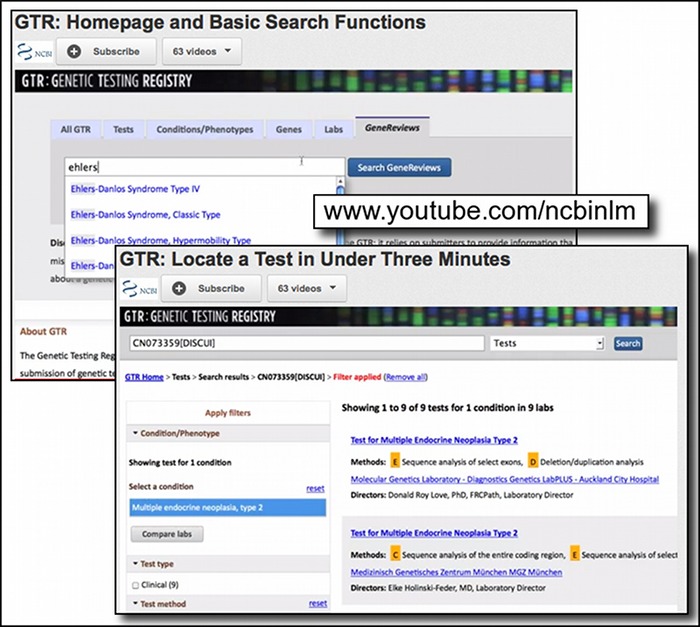
The Genetic Testing Registry: Finding Genetic Tests and Related Information
The NCBI recently released the Genetic Testing Registry (GTR). This new resource is a voluntary registry of genetic tests and laboratories with detailed information about the tests and their providers. The initial scope of GTR includes single gene tests for Mendelian disorders, as well as arrays, panels and pharmacogenetic tests. The registry includes detailed information about the purpose of the test, methodology, analytical and clinical validity, and information on clinical usefulness. GTR provides access to information from the GeneReviews book on the NCBI Bookshelf – peer reviewed descriptions of genetic diseases and information on genetics tests and NCBI molecular databases such as Gene. GTR is a central hub for information about genetic conditions and also provides context-specific links to a variety of resources, including practice guidelines, published literature, and genetic information. As mentioned in the previous section of this newsletter, two new videos on the NCBI YouTube channel provide quick introductions to the GTR. The original NIH press release has more information about the GTR.
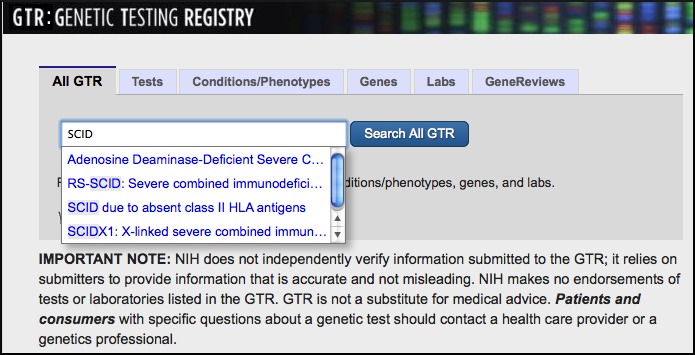
BLAST News
BLAST 2.26+ Release
The latest version of the C++ build of BLAST+ (2.2.26) is now available from the BLAST FTP area and is running on the NCBI BLAST Web service. This new BLAST+ release contains a number of important changes and improvements including the three listed below.
Domain Enhanced Lookup Time Accelerated BLAST (DELTA-BLAST) is a new BLAST algorithm that can be more sensitive than standard protein-protein BLAST. DELTA-BLAST identifies conserved domains in the query sequence using Reverse PSI BLAST and then uses this information to construct a Position Specific Score Matrix (PSSM) then performs a PSSM search against the BLAST protein database. DELTA-BLAST can be invoked on the Protein-protein BLAST Web Service by selecting the DELTA-BLAST radio button in the “Program Selection” area of the submission form. The standalone BLAST package has DELTA-BLAST as a separate program (deltablast). Running DELTA-BLAST locally requires a special version of CDD database (cdd_delta) available from the BLAST db directory on the FTP site.
A new Finite Size Correction has been added to the to the blastp algorithm to improve the accuracy of BLAST statistics (Expect values). The new finite size correction especially improves statistics for matches for short query or short database sequences.
Standalone BLAST now contains the program makeprofiledb, a C++ coded replacement for the NCBI C toolkit program formatrpsdb. Makeprofiledb can generate search sets for RPS-BLAST, including the specialized data needed by DELTA-BLAST.
Final version of C-toolkit BLAST Package
Version 2.2.26 is the final version of NCBI C language toolkit BLAST. The source code for these applications will no longer be developed, but will continue to be available. Users of these legacy programs should migrate to the BLAST+ applications that are being actively developed. The BLAST Command Line Applications User Manual provides help on transitioning to the BLAST+ applications.
Netblast (blastcl3) Service Discontinued: Replaced by remote Option in BLAST+
The Netblast client (blastcl3) that has provided batch search access to the NCBI Web BLAST service will be discontinued in the near future. The BLAST+ applications replace and improve upon the functions provided by blastcl3. Blastcl3 users should switch to BLAST+ as soon as possible. Locally installed BLAST+ applications can perform remote searches using the NCBI Web service when the ‘remote’ option is included on the command line. The BLAST+ remote service has a number of advantages over the blastcl3 application. Blastcl3 requires a persistent connection during the entire search, can only submit one query at a time, and is unable to return the BLAST Request ID (RID) used in the search. The BLAST+ remote service can submit multiple queries (from FASTA input) at once, poll for the results using the BLAST RID, and also print the RID in the BLAST report. Using the BLAST RID, it is possible to reformat the search locally with the blast_formatter application, reformat the search at the NCBI web site, or use analysis tools such as the BLAST treeview or the taxonomy report.
Changes in the BLAST Database List on the NCBI Web Services
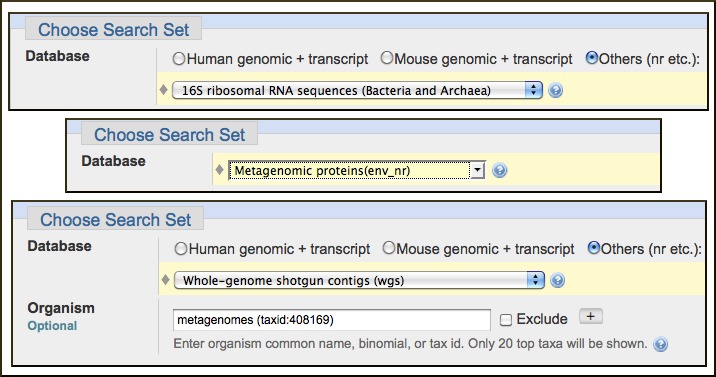
A new microbial 16S ribosomal RNA sequence database is now available on nucleotide-nucleotide BLAST search page. This database contains Archaeal and Bacterial 16S sequences from the Archaeal 16S Ribosomal RNA and Bacterial 16S Ribosomal RNA Targeted Loci Projects. This database should be helpful in classifying unknown microbial 16S sequences from a wide range of sources.
Sequences from environmental samples formerly available in the env_nr and the env_nt databases are now available in the Metagenomic proteins database and, for nucleotide sequences, through the Whole Genome Shotgun Contigs (WGS) database by selecting “metagenomes (taxid: 408169)” as an Organism limit.
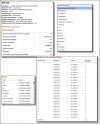
The following image shows the selections needed on the BLAST submission form to search these three new or modified databases.
CDD Results Now Shown for Translated BLAST (blastx) Searches
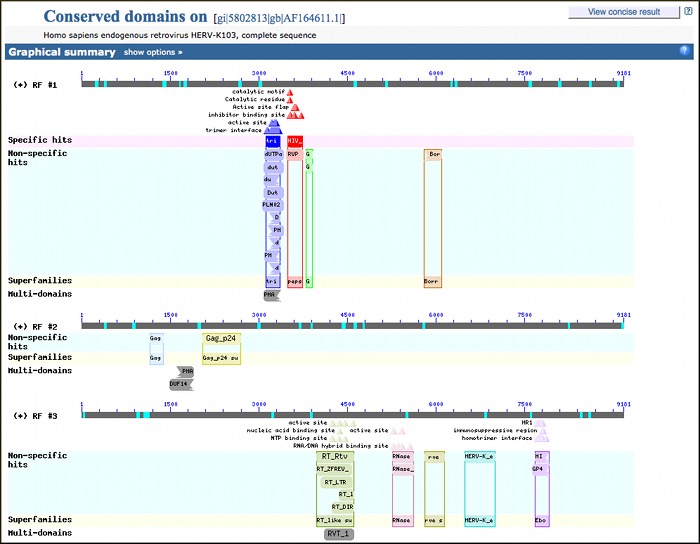
Conserved Domain Search results are now provided for all translated (blastx) searches with query sequences shorter than 10,000 bases. Conserved domain searches are performed with all six reading frames of the query sequence and results are reported for each frame that has matches. This is very useful for helping to characterize coding regions on genomic regions as shown immediately below from the results for a blastx search with a human endogenous retrovirus (AF164611).
Remap and Variation Reporter: Two New Services for Mapping Locations onto Genome Builds
The Genome Remapping Service (Remap) and the Variation Reporter are related tools that find locations on current and past genome builds.
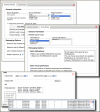
The Remap tool translates or projects the coordinates of genes, variants (SNPs), and other sequence-based markers from one genome assembly (build) to another for human, mouse, rat, zebrafish and sea urchin (Strongylocentrotus purpuratus). It also includes a Clinical Remap version that performs coordinate remapping between genome assemblies and the reference standard RefSeqGene records. Figure 3 and Figure 4 show the submission and results for the Remap service. Locations to be projected can be in a variety of common genome annotation formats such as UCSC Browser Extensible Data (BED) format, Gene Transfer Format (GTF), Generic Feature Format (GFF and GFF3), Human Genome Variation Society (HGVS) nomenclature, and Genome Variation Format (GVF) among others. When projection of features is successful, the service reports the new locations with the submitted annotations in the selected format for downloading and also provides output in a format suitable for loading into Genome Workbench, the NCBI’s standalone sequence analysis and annotation platform. A programming interface (API) is also available for the Remap service. A demonstration PERL script (remap_api.pl) that accesses the service is available from the NCBI FTP site.
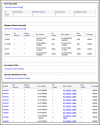
The Variation Reporter, shown in Figure 5, takes a set of locations in a human genome assembly and identifies known human variations (NCBI Reference SNPs) at those positions. This service is particularly helpful for identifying experimentally or clinically determined variants. Like the Remap service, the Variation Reporter accepts a variety of genome annotation formats – HGVS, GVF and BED. The results provide the location of the variants in the selected build and important information about any identified known variants including the dbSNP ID, the known allele, and, if available, clinical information, minor allele frequency, links to literature, and functional consequences. The results also provide the genomic context by displaying the mapped locations in the graphic sequence viewer (Figure 5, bottom panel). The Remap Service and the Variation Reporter are useful for interconverting annotations between genome builds and mapping and identifying experimentally determined variants.
NCBI Aspera Download Site Available for NCBI Databases and Tools
An Aspera protocol download site is available as an alternative to FTP for all NCBI downloads. The Aspera protocol provides a much faster transfer rate and is most important for downloading very large data sets such as those from next-generation sequencing studies, but can be used to improve download performance for any public NCBI data files or software packages. The Aspera protocol site requires the free AsperaConnect client application available from Aspera Connect. The Aspera Transfer Guide, available on the NCBI Bookshelf, provides additional information on using the fast download site.
1000 Genomes Project Data Now on Amazon Cloud Service
As announced in the recent NIH press release, data from the 1000 Genomes project - the world's largest set of data on human genetic variation produced by the international 1000 Genomes Project — are now publicly available on the Amazon Web Services (AWS) cloud. 1000 genomes data may also be downloaded from the NCBI though FTP or through the Aspera protocol site.
Microbial Genomes Update
One hundred ninety nine finished microbial (archaeal and bacterial) genomes were released from November 2011 through March 2012. The original sequence data files submitted to the International Sequence Database Collaboration (INSDC) are available in the Bacteria directory in the genomes area of the GenBank FTP site. RefSeq provisional versions were released for a selected set of 118 of the complete INSDC microbial genomes during the same period. These are available from the /genomes/Bacteria directory on the FTP site.
In addition, data from 1,135 microbial whole genome-shotgun (WGS) sequencing projects were added to the INSDC during this period. The original submitted files are available in the Bacteria_DRAFT directory in the GenBank genomes area. RefSeq provisional versions of 210 WGS microbial projects were released in the /genomes/Bacteria_DRAFT area of the FTP site.
All GenBank and RefSeq microbial genomes are incorporated in the NCBI integrated Entrez search and retrieval system and the BLAST sequence similarity search service.
NCBI Articles in Nucleic Acids Research Database Issue
The Nucleic Acids Research 2011 Database Issue contains 10 articles about NCBI resources, tools, and databases including BioAssay, SRA, GEO, BioProject / BioSample, Taxonomy Epigenomics, MMDB (Structure), RefSeq and GenBank. Free full-text articles from the database issue are available from PubMed Central and the publisher’s site and are linked to the summaries and abstracts in PubMed.
GenBank News
GenBank release 189 is available through Entrez, BLAST and from the GenBank FTP area. The current release incorporates data available as of April 15, 2011 and, with the whole-genome shotgun portion, contains 411,959,832,946 bases from 232,729,719 sequence records. Release notes describe the current state of data and upcoming changes. The GenBank page provides more information on the database content and scope as well as submission information.
RefSeq News
RefSeq Release 52
RefSeq Release 52 is available through Entrez, BLAST, and from the RefSeq FTP area. The current release includes 20.2 million Reference Sequence records from 16,923 different species or strains. The RefSeq release notes provide more detailed information.
RefSeq Genome Annotation Files in GFF3 Format
NCBI now offers Reference Sequence (RefSeq) genome annotation files in the latest Generic Feature Format (GFF3) specification (1.20). RefSeq genome data can be downloaded from the genomes area of the NCBI FTP site. GFF3 files are in the GFF directory within each organism directory. Currently GFF3 files are available for the NCBI annotations of the latest assemblies for human, cow, dog, chicken, and many others.
Keeping Up with NCBI
Seventeen topic-specific mailing lists are available that provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page. Subscribe to the NCBI Announce list to receive updates on the NCBI News.
Twenty-five RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce.
NCBI’s Facebook page and Twitter feed also provide updates on NCBI resources.
Send comments and questions about NCBI resources to info@ncbi.nlm.nih.gov, or call 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI Discovery Workshops May 15-16 at NLM: Seats still available
- Assembly: a Companion to the Genome Database
- New Videos on NCBI’s YouTube Channel
- The Genetic Testing Registry: Finding Genetic Tests and Related Information
- BLAST News
- Remap and Variation Reporter: Two New Services for Mapping Locations onto Genome Builds
- NCBI Aspera Download Site Available for NCBI Databases and Tools
- 1000 Genomes Project Data Now on Amazon Cloud Service
- Microbial Genomes Update
- NCBI Articles in Nucleic Acids Research Database Issue
- GenBank News
- RefSeq News
- Keeping Up with NCBI
- NCBI News, April 2012 - NCBI NewsNCBI News, April 2012 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...