NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
New version of Genome Workbench is Available
Monday, November 19, 2012
Version 2.6.5 of Genome Workbench has been released with some new features and many improvements and bug fixes.
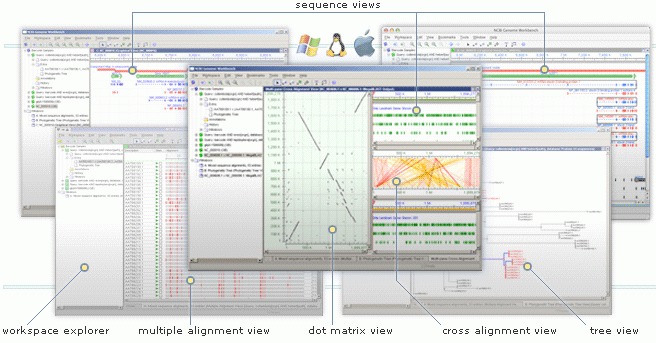
From the Genome Workbench Release Notes:
New Features
- Add Tools Quick Launch items to context menu
- Graphical Sequence View: implemented separation graphs into sub-tracks based on meta-information title
Bug Fixes and Improvements
- Updated VCF and GFF readers, fixed issues with checking sequence ids
- Connection issues resolved for users running international versions of Windows or MacOS
- BLAST tool dialog freeze for a long time loading list of available BLAST databases
- BLAST RID load: fixed to automatically import both alignments and local sequences
- MUSCLE tool integration: fixed alignment coordinate remapping(shift error)
- Graphical Sequence View: fixed display of tracks with alt-loci alignments
- Graphical Sequence View: fixed fixed alignment score coloration for SRA
- Graphical Sequence View: tooltips improved alignment statistics and positions
- Graphical Sequence View: fixed indels rendering in protein-to-genomic alignments
- Graphical Sequence View: fixed empty tooltip lines (sequence)
- Multiple Alignment View: fixed tooltips to be displayed only over sequence part
- Linux: improved README for users building custom Genome Workbench
- Search View: fixed failures handling multiple sequence ids
- Search View: Kozak patterns updated
- BAM/cSRA support: fixed Open File masking for MacOS and Linux
- Text View: fixed crash in Flat File mode
- Tree View: fixed view title after tree sort operation
- MacOS: fixed opening projects from network paths/mappings/mountpoints
- MacOS: fixed issues with pasting into Open View dialog
- HTTP connection component: fixed "User-Agent:" self-identification to better conform to standards
- Improved logging and Feedback reporting not to collect potentially sensitive information
RefSeq Release 56 is Available for FTP
Wednesday, November 14, 2012
The complete RefSeq release 56 contains 23,892,460 records including 2,729,041 genomic sequences, 3,185,652 RNAs, and 17,977,767 proteins from 18,512 different organisms.
See the Release statistics file or Release notes for more information.
Please note that this update includes information from dbSNP Build 137, and includes incremental updates for human records.
In addition, in the first quarter of 2013 the bacterial RefSeq collection will expand to include more genomes that represent complete or draft assemblies from novel microbial isolates as well as clinical and population samples. As part of this expansion, bacterial RefSeq genomes will be re-annotated to increase consistency across this dataset.
New CDD release Available & now Mirrors TIGRFAM v13
Monday, November 05, 2012
Version 3.09 of the Conserved Domain Database (CDD) is now available. 46,629 conserved domain models are indexed for searching at NCBI and include a mirror of TIGRFAM v13.
The new data are incorporated in the CDD website and Conserved Domain BLAST Search services. More detailed statistics are available from the CDD News page. CDD matrices and other information can be downloaded from the FTP site.
NCBI will be Presenting and Exhibiting at ASHG 2012
Monday, November 05, 2012
NCBI Staff members will present, display posters, run a workshop and exhibit at the ASHG 2012 Annual Meeting in the Moscone Center, San Francisco, CA which takes place from Tuesday, November 6th through Saturday, November 10th, 2012.
Wednesday, November 7 through Friday, November 9, 2012 from 10am-4:30pm:
- Come visit us at the NCBI Booth (#224) in the Exhibit Hall, Lower Level South
- NCBI staff will be available to answer questions, listen to your suggestions, and offer live demonstrations with NCBI tools and databases.
Presentations on Tuesday, November 6, 2012
- 9-9:15am: "Managing information about human phenotype at NCBI" in the "Getting Ready for The Human Phenome Project" Satellite Meeting - San Francisco Marriott Marquis, Section 7, Yerba Buena Ballroom, Lower Level B2
- 1-4pm: "Getting the Most from the Human Genome: Understanding Updates and Improvements in the Reference Assembly" in the "Genome Reference Consortium Workshop" Moscone Center, Room: 236/238, East Mezzanine Level South
Workshop on Wednesday, November 7, 2012
- 12:15-2:15pm: " Workshop: Discovering Biological Data at NCBI" in the Moscone Center, Room: 304/306, Esplanade Level South
- This workshop provides an introduction to using the Entrez system to perform searches and find related data starting with a list of reviewed human genes. Specific tasks covered include finding reference sequences, mapping variations, identifying homologous genes, exploring expression studies, and using MyNCBI to save searches and manage data.
Presentations on Saturday, November 10, 2012
- 9:40-11:40am: " Improving the accuracy of variant identification" & "Introducing ClinVar" in the "Centralizing the Deposition and Curation of Human Mutations" Section, Moscone Center, Room: 132, Lower Level North
For the full list of NCBI's events, see the "NCBI at ASHG 2012" Schedule page.
- NCBI News, November 2012 - NCBI NewsNCBI News, November 2012 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
