NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
BLAST+ 2.2.30 released
Thursday, October 30, 2014
A new version (2.2.30) of the stand-alone BLAST executables is now available, bringing several improvements to BLAST+. These improvements include tasks for BLASTX and TBLASTN (blastx-fast and tblastn-fast) that use longer words, as described in Shiryev, Papadopoulos, Schaffer, and Agarwala (2007), as well as support for composition-based statistics in RPS-BLAST. A number of bug fixes, including those for FASTA parsing, are also included.
The tarballs/installers are located on the FTP site. LINUX, Windows, and MacOSX executables are available here. The BLAST AMI at AWS will also be updated to 2.2.30 (see this BLAST Help page for information).
For more information, please see the full release notes.
New Genome BLAST selector on the BLAST homepage
Tuesday, October 28, 2014
You can now easily find Genome-specific BLAST pages using the search box on the BLAST homepage under the “BLAST Assembled Genomes” section. This new feature allows you to quickly access and search BLAST databases for the genome of an organism of interest.
Figure
Figure 1. The genome BLAST autocomplete on the BLAST homepage (top right). The autocomplete provides matches to organism (common and scientific binomials) and strain names to find genomic datasets.
Simply start typing your organism name into the box and suggestions will appear. The autocomplete accepts species or strain-level eukaryotic and microbial names as well as metagenomic taxa (community and organism associated metagenomes).
Once you select a suggestion, you will be taken to a BLAST page with the best (most complete, reference) genomic database preselected. In cases where there is no assembled genome sequence, the page will load with whole genome shotgun databases for the organism. If there is no specific genome-sequencing project, the page will load with default nucleotide database (nr/nt) limited to the organism of interest.
Next NCBI webinar on November 5th
Thursday, October 23, 2014
On November 5th, NCBI will have a webinar entitled “Exploring and Downloading Sequences and Annotations for Genomes and Metagenomes at the NCBI”. This presentation will introduce you to how NCBI processes genome-level data and produces annotation through the prokaryotic and eukaryotic genome annotation pipelines and show you how to access and download these data from the NCBI site.
You will learn to find, browse, and download genome-level data for your organism of interest and for environmental and organismal metagenomes using the BioProject and Assembly resources. In addition to assembled and annotated data, you will see how to retrieve and download draft whole genome shotgun and read-level next-gen sequencing data from the Nucleotide and Sequence Read Archive (SRA) databases. You will also see how to access results of precomputed analyses of genomes, as well as perform your own analyses of assembled and unassembled genomic data using NCBI's genome BLAST and SRA-BLAST services.
To register: https://attendee.gotowebinar.com/register/7154056329796392706
See materials and video from previous webinars and descriptions of upcoming webinars on the NCBI Webinars page.
GenBank release 204.0 is now available via FTP
Wednesday, October 22, 2014
Release 204.0 (10/20/2014) has 178,322,253 non-WGS, non-CON records containing 181,563,676,918 base pairs of sequence data. In addition, there are 196,049,974 WGS records containing 805,549,167,708 base pairs of sequence data.
During the 63 days between the close dates for GenBank Releases 203.0 and 204.0, the non-WGS/non-CON portion of GenBank grew by 15,840,696,543 base pairs and by 4,213,503 sequence records. During that same period, 532,480 records were updated; an average of 75,333 non-WGS/non-CON records were added and/or updated per day. Between releases 203.0 and 204.0, the WGS component of GenBank grew by 31,497,068,977 base pairs and by 6,969,555 sequence records.
The total number of sequence data files increased by 123 with this release. The divisions are as follows:
- BCT: 10 new files, now a total of 152
- CON: 8 new files, now a total of 286
- ENV: 2 new files, now a total of 76
- EST: 1 new file, now a total of 477
- INV: 84 new files, now a total of 124
- PAT: 4 new files, now a total of 214
- PLN: 3 new files, now a total of 89
- VRT: 11 new files, now a total of 44
For downloading purposes, please keep in mind that the uncompressed GenBank flatfiles are approximately 680 GB (sequence files only). The ASN.1 data require approximately 557 GB.
More information about GenBank Release 204.0, including important changes included in this release, is available in the release notes.
dbSNP human Build 142 released
Friday, October 17, 2014
dbSNP human Build 142, based on the GRCh38 and GRCh37.p13 assemblies, is now available on the integrated NCBI Entrez system and through FTP. Build 142 provides 112 million Reference SNP (RS) clusters, including 51 million new RS created from 1000 Genomes Phase III variants as well as from other large sequencing projects. To see complete build statistics, visit the dbSNP summary page. For more information on Build 142, please see this dbSNP listserv announcement.
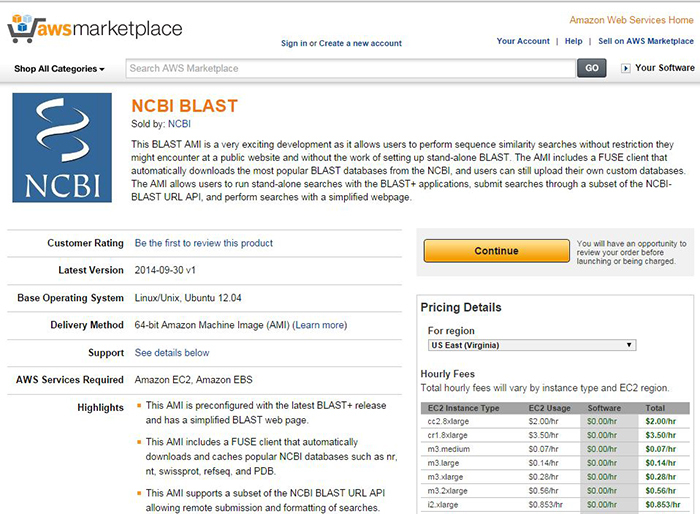
Amazon Web Services (AWS) Marketplace provides the easiest way to start an NCBI BLAST instance
Thursday, October 16, 2014
BLAST instances can now be started from the Amazon Web Services (AWS) Marketplace. Using the Marketplace is the easiest way to start a BLAST instance at AWS. In addition, users who subscribe to the BLAST package will be notified when it is updated.
Figure 1. NCBI BLAST on the AWS Marketplace.
As reported this summer, the BLAST instance at AWS is packaged as an AMI (Amazon Machine Image), which allows users to run stand-alone searches with the BLAST+ applications, submit searches through a subset of the NCBI-BLAST URL API, and perform searches with a simplified web page. The BLAST AMI also includes a FUSE client that can download BLAST databases during the first search.
Variation Reporter version 1.4 released
Wednesday, October 15, 2014
Variation Reporter has just been updated to version 1.4 and now supports querying and reporting on both GRCh37 and GRCH38. For more information about other improvements, see the Variation Reporter release notes.
Variation Reporter is NCBI’s tool for matching user-uploaded variant locations against known dbSNP, dbVar and ClinVar data.
Conserved Domain Database (CDD) version 3.12
Tuesday, October 14, 2014
Conserved Domain Database (CDD) version 3.12 is now available with 1526 new or updated NCBI-curated domains and 49,955 total domain models from CDD’s database providers: Pfam, SMART, COG, TIGRFAMs, Protein Clusters, and the NCBI in-house curation project.
You can access CDD at the Conserved Domains homepage and find updated content on the CDD FTP site.
Updates to assembly alignments for NCBI Remap service
Friday, October 10, 2014
NCBI has updated its assembly alignment software (now version 1.7), which generates the alignments used for Remap, NCBI's coordinate remapping service. The improvements include: better handling of alternate loci and fix patches, improved alignments in regions of copy number variation, better recognition of sequence regions that are unaltered between two versions of a WGS assembly, incorporation of fixes to BLAST including bugs affecting alignments around regions with multiple mismatches and indels, and assorted other quality improvements.
These changes improve coordinate remapping from one assembly to another, resulting in better accuracy when remapping features like SNPs or gene annotation to their inferred locations on a new assembly.
Most of the alignment sets available from the Remap service have been regenerated with the v1.7 software, including human GRCh37.p13 x GRCh38. Alignments are available for remapping between various assemblies for 36 taxa, including:
- human GRCh38 (GCF_000001405.26)
- mouse GRCm38.p3 (GCF_000001635.23)
- rat Rnor_6.0 (GCF_000001895.5)
- zebrafish GRCz10 (GCF_000002035.5)
Remap can be used through the web interface and a public API, and the assembly alignments are also available in GFF3 format from the remap FTP site.
New NCBI Insights blog: Sequence updates in human assembly GRCh38: improving gene annotation
Thursday, October 09, 2014
The latest blog post on the NCBI Insights blog continues the discussion of GRCh38. This time, the blog post focuses on how GRCh38 improved gene annotation.
Zebrafish (Danio rerio) GRCz10 now annotated
Wednesday, October 08, 2014
Zebrafish (Danio rerio) GRCz10 is annotated! GRCz10 is an update to the Zv9 assembly, released by the Genome Reference Consortium (GRC), which now manages this reference genome assembly in addition to those for human and mouse. GRCz10 includes more than 1,000 new clone sequences and improvements to the order and orientation of assembly sequences.
RefSeq annotation of GRCz10 was produced by the Eukaryotic Genome Annotation Pipeline. It is available in NCBI's sequence and BLAST databases, in Gene, and is ready for download. Gene displays annotation data for both Zv9 and GRCz10 to help transition to the new assembly.
GRCz10 annotation is based on transcript and protein evidence including alignments of nearly 2.3 billion RNA-Seq reads (169 billion bases) from 27 distinct BioSample accessions. A total of 30,741 genes and 63,217 transcripts were identified on GRCz10. This includes 14,442 genes (14,019 protein-coding) with known RefSeq transcripts (NM, NP, or NR accessions), and an additional 16,158 predicted genes (12,459 protein-coding) with model RefSeqs (XM, XP, or XR accessions). Note that predicted genes and model RefSeqs aren't included in some resources outside of NCBI. Detailed statistics of annotation results and input reagents are available in the annotation report. NCBI has also annotated 16 other fish genomes, summarized in the annotated genomes report.
For more information about the updates in GRCz10, please see the GRC zebrafish page or the GRC blog post on the new assembly. See other organisms that were recently annotated or are currently in the annotation pipeline on the Eukaryotic Genome Annotation Pipeline status page.
dbVar now accepts VCF submissions of structural variation data
Friday, October 03, 2014
dbVar, NCBI's database of genomic structural variation, now accepts submissions in the Variant Call Format (VCF) in addition to their other standard formats: Excel, Tab, and XML. Instructions for submitting in VCF can be found on the dbVar Home, Submission Guidelines, and Submission Templates pages.
dbVar VCF requirements are largely identical to those of the standard 1000 Genomes VCF spec v4.1. However, a few minor changes have been made and are detailed in the documentation. VCF files must be accompanied by one or more files containing metadata using one of the standard formats listed above.
For more information, please visit the dbVar homepage.
New NCBI Insights blog post: NCBI's medical genetics resources
Thursday, October 02, 2014
The latest blog post on NCBI Insights, “NCBI’s 3 Newest Medical Genetics Resources: GTR, MedGen and ClinVar”, gives an overview of NCBI’s three medical genetics resources and outlines their content features. A more in-depth introduction is available in the Medical Genetics Resources webinar from June 2014.
NCBI webinar on E-Utilities October 15th
Wednesday, October 01, 2014
On October 15th, NCBI will have a webinar entitled “An Introduction to NCBI’s E-Utilities, an NCBI API.” E-Utilities is a tool to assist programmers in accessing, searching and retrieving a wide variety of data from NCBI servers.
This presentation will introduce you to the Entrez Programming Utilities (E-Utilities), the public API for the NCBI Entrez system that includes 40 databases such as Pubmed, PMC, Gene, Genome, GEO and dbSNP. After covering the basic functions and URL syntax of the E-utilities, we will then demonstrate these functions using Entrez Direct, a set of UNIX command line programs that allow you to incorporate E-utility calls easily into simple shell scripts.
Click here to register.
- BLAST+ 2.2.30 released
- New Genome BLAST selector on the BLAST homepage
- Next NCBI webinar on November 5th
- GenBank release 204.0 is now available via FTP
- dbSNP human Build 142 released
- Amazon Web Services (AWS) Marketplace provides the easiest way to start an NCBI BLAST instance
- Variation Reporter version 1.4 released
- Conserved Domain Database (CDD) version 3.12
- Updates to assembly alignments for NCBI Remap service
- New NCBI Insights blog: Sequence updates in human assembly GRCh38: improving gene annotation
- Zebrafish (Danio rerio) GRCz10 now annotated
- dbVar now accepts VCF submissions of structural variation data
- New NCBI Insights blog post: NCBI's medical genetics resources
- NCBI webinar on E-Utilities October 15th
- NCBI News, October 2014 - NCBI NewsNCBI News, October 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...