NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Updated human and mouse genome annotations now available
Tuesday, March 31, 2015
Updated annotations for the human and mouse RefSeq genomes produced by the Eukaryotic Genome Annotation Pipeline are now available. New known RefSeq transcripts (NM_ and NR_ accessions) and non-transcribed pseudogenes (NG_ accessions) were used for these annotations. The number of model RefSeq predictions (XM_ and XR_ accessions) also increased through the use of additional RNA-Seq datasets, especially for human where model RefSeq annotated on GRCh38.p2 contain 41% more exonic bases (31 MBp) than the known RefSeq.
Homo sapiens annotation release 107: see in Gene, BLAST or download.
- Assemblies annotated: GRCh38.p2 (GCF_000001405.28, reference) and CHM1_1.1 (GCF_000306695.2); note that we removed the HuRef assembly, GCF_000002125.1, from the RefSeq collection.
- Annotation changes for GRCh38.p2:
- 50% more genes with alternative splice variants (an average of 3.52 transcripts per gene)
- 100% more non-coding genes, 146% more non-coding transcripts
- 8% more annotated known RefSeq
Mus musculus annotation release 105: See in Gene, BLAST or download.
- RNA-Seq datasets used: mouse ENCODE transcriptome (PRJNA66167) and a whole-embryo project (PRJNA203332)
- Annotation changes for GRCm38.p3:
- 3.4% more genes with alternative splice variants (an average of 2.87 transcripts per gene)
- 16% more non-coding genes, 27% more non-coding transcripts
- 5.8% more annotated known RefSeq
You can find the annotation runs currently in progress on the Eukaryotic Genome Annotation Pipeline status page.
April 8th webinar: "The NCBI Minute: Introducing MOLE-BLAST"
Wednesday, March 25, 2015
On April 8th, NCBI will present a five-minute webinar introducing MOLE-BLAST, a tool for clustering targeted sequences, like those from 16s rRNA, with database sequences and providing taxonomic context. MOLE-BLAST can quickly establish taxonomy for sequences from uncultured or environmental sequences. To register, click here.
The NCBI Minute is a series of short webinars that give a brief introduction to an NCBI tool or service, as well as quick tips on using our resources. To see upcoming webinars, as well as summaries, recordings (via YouTube) and related materials from past webinars, please see the NCBI Webinars page.
April 1st webinar: "A Practical Guide to Using NCBI BLAST on the Web"
Tuesday, March 24, 2015
Next Wednesday, April 1st, NCBI will present a webinar on the NCBI BLAST service. The webinar will highlight important features and demonstrate the practical aspects of using NCBI BLAST, the most popular sequence similarity service in the world. To register, click here.
Some of the useful features that will be discussed include:
- Access from the Entrez sequence databases
- The new genome BLAST service quick finder
- The integration and expansion of Align-2-Sequences
- Organism limits and other filters
- Reorganized databases
- Formatting and downloading options
- TreeView displays
We will also show you how to use other important sequence analysis services associated with BLAST including Primer-BLAST, iGBLAST, and MOLE-BLAST, a new tool for clustering and providing taxonomic content for targeted loci sequences (16S, ITS, 28S). These aspects of BLAST provide easier access and results that are more comprehensive and easier to interpret.
To see upcoming webinars, as well as summaries, recordings (via YouTube) and related materials from past webinars, please see the NCBI Webinars page.
Update: We will also have a short webinar on MOLE-BLAST on April 8th. Click here to learn more about it and sign up.
dbSNP Build 143 Phase II now available
Tuesday, March 17, 2015
dbSNP build 143 phase II includes data for cow, Ciona intestinalis and prairie vole. Build 143 provides more than 537 million submitted and 299 million reference variants for 9 species. You can access build 143 SNP data through the integrated NCBI Entrez system and through FTP. To see complete build statistics, visit the SNP summary page.
New NCBI Insights blog post: "Exploring Entrez Direct: Parsing the XML Output of E-utilities"
Friday, March 13, 2015
The latest blog post on NCBI Insights shows you how to use Entrez Direct's ability to parse and reformat complex XML data returns from EFetch, using PubMed records as an example.
NCBI homepage update includes action buttons, category pages
Thursday, March 12, 2015
The NCBI homepage now has six new buttons on it: Submit, Download, Learn, Develop, Analyze, and Research. Each of these buttons leads to an action page devoted to a particular set of services.
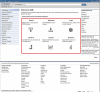
Figure
Figure 1. The NCBI homepage. The new action buttons are outlined in red.
These action pages will allow you to easily access the pages and resources you need to complete tasks. For instance, you can:
- find information about the Entrez API,
- find an upcoming NCBI webinar,
- find an NCBI tool that designs PCR primers,
and much more.
We've also included a blue Feedback button on the left side of the Download, Learn, Develop and Analyze pages so that you can tell us what you think. We look forward to hearing your comments.
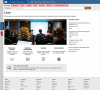
Figure
Figure 2. The Learn page. The six category pages are linked at the top, in the header. On the left side of the page, an arrow points to the feedback tab, which you can use to comment.
On the new action pages, you'll also see 6 categories in the header: Literature, Health, Genomes, Genes, Proteins, and Chemicals. These category pages highlight useful databases, tools and resources for each of the topics all in one place. If you follow us on LinkedIn, these categories will be familiar to you - we've used them as Showcase Pages to group our news stories and announcements by topic.
Stay tuned to NCBI News and to our blog, NCBI Insights, for more information about the new homepage.
NCBI Sequence Viewer version 3.6 available
Wednesday, March 11, 2015
NCBI Sequence Viewer has recently been updated and now has improved rendering of SNP insertions/deletions and narrow features, as well as better graph track names. A full list of new features, improvements and fixes is included in the release notes.
Sequence Viewer is a graphical view of sequence sand color-coded annotations on regions of sequences stored in the Nucleotide and Protein databases.
March 18th webinar: "Using the dbGaP Data Browser to browse aligned reads and genotypes from the Database of Genotypes and Phenotypes"
Tuesday, March 03, 2015
In two weeks, NCBI will present a webinar on the dbGaP Data Browser. This webinar will show you how to use the Data Browser to access aligned reads and genotypes, using the last exon of the APOE gene from an Alzheimer's disease study as an example. To register, click here.
The dbGaP Data Browser provides access to aligned reads and genotypes from a variety of sequencing studies from the Database of Genotypes and Phenotypes (dbGaP). The browser shows sample-level alignments - in the context of the genome sequence - with variants from dbSNP and known clinical variants such as those from ClinVar, as well as differences from the reference genome sequence. The browser allows you to filter the subjects by a variety of indexable values and, depending on your level of access, view-only or downloadable access to reads and genotypes.
To see upcoming webinars, as well as summaries, recordings via YouTube, and related materials from past webinars, please see the NCBI Webinars page.
- Updated human and mouse genome annotations now available
- April 8th webinar: "The NCBI Minute: Introducing MOLE-BLAST"
- April 1st webinar: "A Practical Guide to Using NCBI BLAST on the Web"
- dbSNP Build 143 Phase II now available
- New NCBI Insights blog post: "Exploring Entrez Direct: Parsing the XML Output of E-utilities"
- NCBI homepage update includes action buttons, category pages
- NCBI Sequence Viewer version 3.6 available
- March 18th webinar: "Using the dbGaP Data Browser to browse aligned reads and genotypes from the Database of Genotypes and Phenotypes"
- NCBI News, March 2015 - NCBI NewsNCBI News, March 2015 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
