NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Epigenomics database to be retired June 1, 2016
Friday, May 27, 2016
The Epigenomics database, a public repository that was developed to archive genome-wide maps of DNA and histone modifications, will be retired on June 1, 2016.
All epigenomics data are available in our GEO resource. If you are specifically interested in the NIH Roadmap Epigenomics Project, we will maintain a page for this project's data.
NCBI launches new Twitter account for NCBI Bookshelf
Monday, May 23, 2016
NCBI has a new Twitter feed - @ncbibooks - to announce new books and documents available on the NCBI Bookshelf. An online resource providing free access to the full text of books and documents in life sciences and health care, the Bookshelf currently provides access to over 4,500 titles.
The Bookshelf is continuously expanding with new materials as well as receiving updates to existing books & documents. Between May 16, 2016 and May 20, for example, 19 new titles were added. Among the new titles are several Agency for Healthcare Research and Quality reports (for example, a comparative effectiveness report on imaging for pretreatment staging of small cell lung cancer), health technology assessments and systematic reviews from Canadian Agency for Drugs and Technologies in Health, and National Institute for Health Research (UK), and World Health Organization guidelines on daily iron supplementation.
Keep on top of the newest releases by following us on Twitter at @ncbibooks!
For general NCBI news, follow us on Twitter, Facebook and LinkedIn.
New NCBI Insights blog post: Fast Sequence Inspection with ORFfinder and SmartBLAST (PubMed Labs)
Monday, May 16, 2016
The latest blog post on NCBI Insights describes the latest PubMed Labs experiment, ORFfinder. ORFfinder is a graphical analysis tool for reading open reading frames (ORFs). See NCBI Insights to read about the new features and leave feedback. We look forward to hearing your thoughts on ORFfinder.
PubMed Labs is an initiative for creating innovative and relevant products by involving you, our user community, from the beginning. It is centered upon our user community, experimentation, learning, and conversation.

RefSeq release 76 is now available
Monday, May 16, 2016
RefSeq release 76 is accessible online, via FTP and through NCBI’s programming utilities. This full release incorporates genomic, transcript and protein data available as of May 9, 2016 and includes 97,792,976 records, 63,971,766 proteins, 14,965,826 RNAs, and sequences from 59,995 organisms. The release is provided in several directories as a complete dataset and also as divided by logical groupings.
More information about release 76 can be found in the release notes. For more information about the RefSeq project, please see the RefSeq homepage.
Genome Browsers section added to Gene
Monday, May 16, 2016
Gene pages now have a new link section in the sidebar called "Genome Browsers". This section provides an easy way to access all of your favorite browsers, including:
- The NEW Genome Data Viewer, available for over 300 species
Go to the BRCA1 page or search for your favorite gene to try out the links available.
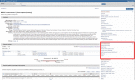
Figure
Figure 1. The Genome Browsers section is on the right side of Gene record pages (outlined in red).
Gene integrates information from a wide range of species. A gene record may include nomenclature, RefSeqs, maps, pathways, variations, phenotypes, and links to genome-, phenotype-, and locus-specific resources worldwide.
Sequence Viewer version 3.14 upgrades platform
Friday, May 13, 2016
Sequence Viewer 3.14, now available, upgrades the platform to use ExtJS 5. Embedders may need to review their pages and make adjustments to accommodate new APIs.
For a full list of features, improvements and bug fixes, see the release notes.
Sequence Viewer is a graphical view of sequences and color-coded annotations on regions of sequences stored in the Nucleotide and Protein databases.
New NCBI Variation summary page highlights all organisms in dbSNP or dbVar with full assembly annotations
Friday, May 13, 2016
The NCBI Variation summary page lists all available organisms in dbSNP and/or dbVar with full assembly annotations. On this new page, you can quickly find out the types and status of genetic variation data for each organism, as well as links to that data. The list is updated regularly with each dbSNP and dbVar new release.
NCBI launches web-based iCn3D, a new viewer for 3D macromolecular structures
Friday, May 13, 2016
NCBI has created iCn3D 1.0, a new WebGL-based viewer for interactive viewing of 3D macromolecular structures and chemicals on the web. Users no longer need to install a separate application to view structures. With iCn3D 1.0, users can:
- Interactively view 3D structures and corresponding sequence data,
- Interactively view superpositions of similar structures,
- Customize the display of a structure and generate a URL that allows you to share the link,
- And incorporate iCn3D into your own pages.
iCn3D can be accessed from the molecular graphic that appears on the structure summary for any record in the Molecular Modeling Database (MMDB).
The source code for iCn3D is available from GitHub for developers who would like to customize the program and/or contribute code, and for users who would like to run the program on their local computer.
For those who still would like to use Cn3D, the executable program version of the 3D structure viewer, it is still available; you can still access structures in the "Download Structure Data" portlet by selecting "Download: ASN.1(Cn3D)".
NCBI annotates 300th organism with the Eukaryotic Genome Annotation Pipeline
Thursday, May 12, 2016
The NCBI Eukaryotic Genome Annotation Pipeline celebrates the annotation of its 300th organism this month! The lucky 300th is Sinocyclocheilus anshuiensis, a cavefish of interest for its adaptation to subterranean habitats. Vertebrates represent about two thirds of the list of 300, but invertebrates and higher plants are also represented. Recently, NCBI has annotated the rice (Oryza sativa subspecies Japonica) and the tobacco (Nicotiana tabacum) genomes.
Data produced by the Eukaryotic Genome Annotation Pipeline is available in the Reference Sequences (RefSeq) collection, BLAST non-redundant and organism-specific databases, Gene database, and is downloadable from the NCBI FTP site. See the full list of annotated organisms, and request the annotation of your favorite!
Genome Workbench 2.10.5 now available
Wednesday, May 11, 2016
Genome Workbench 2.10.5 brings a number of new features, improvements and fixes including ProSplign integration, updates to Tree View, and new export functionalities. For the full list of changes, see the release notes.
Preview the new BLAST home page!
Tuesday, May 10, 2016
NCBI has released a new home page for BLAST. This new page is available through a link on the current home page. The current home page will be replaced by the new page on June 9, 2016.
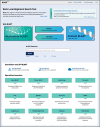
Figure
Figure 1. The new BLAST homepage.
The new design provides improved navigation, a cleaner look, and easier access to new BLAST services, such as running BLAST on the cloud.
Please take a moment to preview the new design. If you have comments or suggestions, please write to vog.hin.mln.ibcn@pleh-tsalb.
NCBI and RCSB PDB to assist ISCB in Sequence-Structure hackathon at ISMB Orlando 2016
Tuesday, May 10, 2016
From the evening of July 8th to the morning of July 11th, NCBI and the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) will assist the International Society for Computational Biology (ISCB) in hosting a sequence-structure hackathon focused on integrating protein structure information and viewers with genomic sequence and variants, pharmacogenomic binding, and general laboratory practice. To apply for this hackathon, complete this form (approximately 10 minutes to complete). Applications are due June 1st, 2016 by 5 pm ET.
This event is for students, postdocs and investigators or other researchers already engaged in the use of JavaScript-based viewers for protein structure visualization and/or genome/gene/protein sequence browsers. It is open to anyone selected for the hackathon, and registered for the ISMB 2016 meeting.
Update: Due to the many potential conflicts for ISMB participants, we have moved all hackathon sessions to the evening, after programming.
Working groups of 5-6 individuals will be formed into five or six teams. These teams will build pipelines and tools to analyze large datasets within a cloud infrastructure. The potential subjects for this iteration are:
- Presenting data to and from JavaScript-viewable protein structures,
- Simplified structure diagrams,
- A digital notebook for structural biologists,
- And pharmacogenomic association in conserved binding sites*.
* Some of the projects will build on APIs feeding data to or extracting data from viewers, so applicants need not be working on JavaScript viewers per se. For example, there is an opportunity to interface existing applications and libraries using any programming language with the Macromolecule Transmission Format (MMTF) for ultrafast access, parsing, and processing of PDB structures.
Please see the application for specific team projects.
Organization
After a brief organizational session, teams will spend three days analyzing a challenging set of scientific problems related to a group of datasets. Participants will analyze and combine datasets in order to work on these problems.
Datasets
Datasets will come from the public repositories housed at the NCBI, PDB and elsewhere.
Products
All pipelines and other scripts, software and programs generated in this course will be added to a public GitHub repository designed for that purpose. A manuscript outlining the design and usage of the software tools constructed by each team will be submitted to an appropriate journal.
Application
To apply, complete this form (approximately 10 minutes to complete). Applications are due June 1st, 2016 by 5 pm ET. Participants will be selected from a pool of applicants based on the experience and motivation they provide on the form. Prior participants and applicants are especially encouraged to reapply. The first round of accepted applicants will be notified on June 3rd by 5 pm ET, and have until June 6th at noon to confirm their participation.
If you confirm, please make sure it is highly likely you can attend, as confirming and not attending bars other data scientists from attending this event. We understand this hackathon is going on during an information-dense scientific meeting, so we ask that attendees attend at least part of three out of five scientific sessions in the draft daily schedule. Please include a monitored email address, in case there are follow-up questions.
Note: Participants will need to bring their own laptop to this program. A working knowledge of scripting (e.g., Shell, Python) is necessary to be successful in this event. Knowledge of JavaScript will be particularly helpful in this event. Applicants must be willing to commit to all three days of the event. No financial support for travel, lodging or meals is available for this event. Also note that the event may extend into the evening hours on Friday and/or Saturday. Please make any necessary arrangements to accommodate this possibility. Awards for achievements like “Most Prolific Code Development” will be given at the ISCB awards ceremony on Tuesday.
Please contact vog.hin@ybsub.neb with any questions.
May 18th webinar: Using VDB BLAST Clients to Search Whole Genome Shotgun Contigs (WGS) and Transcriptome Shotgun Assembly (TSA) Data at the NCBI
Tuesday, May 10, 2016
On May 18th, NCBI will present a webinar that will show attendees how to use the standalone VDB blast programs (blastn_vdb and tblastn_vdb), which are part of the SRA Toolkit, as clients to search whole genome shotgun contigs (WGS) and Transcriptome Shotgun Assembly (TSA) data. WGS, which are partially assembled genome sequences, and TSA, which are transcripts assembled from next-gen RNA-Seq data, are two of the fastest growing categories of sequence data available for BLAST searching.
Through this webinar, you will see how to use the WGS/TSA browser or the EDirect utilities to find the WGS and TSA projects that interest you. You'll also learn how to use an NCBI Perl script to retrieve the database prefixes for taxonomic subsets of WGS databases.
Time and date: May 18th, 2016 12PM Eastern
Registration URL: http://bit.ly/24Ipwu0
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
New NCBI video on YouTube: ProSplign comes to Genome Workbench
Friday, May 06, 2016
The newest video on the NCBI YouTube channel, Genome Workbench: Use ProSplign for Protein to Genomic Alignments, shows you how to use ProSplign within Genome Workbench.
ProSplign is a global alignment tool that produces accurate spliced alignments and locates alignments of distantly related proteins with low similarity.
Subscribe to the NCBI YouTube channel to receive alerts about new videos ranging from quick tips to full webinar presentations.
New NCBI video on YouTube: Submitting BioSample Data to NCBI
Tuesday, May 03, 2016
The newest video on the NCBI YouTube channel, Submitting BioSample Data to NCBI, gives you tips to make the BioSample portion of the data submission process easier.
A BioSample is a description of the biological source materials used in experimental assays.
Subscribe to the NCBI YouTube channel to receive alerts about new videos ranging from quick tips to full webinar presentations.
GenBank release 213.0 is now available via FTP
Monday, May 02, 2016
GenBank release 213.0 (04/14/2016) has 193,739,511 "traditional" (non-WGS, non-CON) records containing 211,423,912,047 base pairs of sequence data. In addition, there are 338,922,537 WGS records containing 1,452,207,704,949 base pairs of sequence data, as well as 98,147,566 TSA records containing 87,811,163,676 base pairs of sequence data.
During the 61 days between the close dates for GenBank releases 212.0 and 213.0, the traditional portion of GenBank grew by 4,405,715,980 base pairs and by 3,489,276 sequence records. During that same period, 539,559 records were updated. An average of 66,046 traditional records were added and/or updated per day.
Between releases 212.0 and 213.0, the WGS component of GenBank grew by 52,342,209,341 base pairs and by 5,909,777 sequence records; during the same period, the TSA component of GenBank grew by 5,878,608,582 base pairs and by 6,015,248 sequence records.
The total number of sequence data files increased by 29 with this release. The divisions are as follows:
- BCT: 14 new files, now a total of 238
- CON: 4 new files, now a total of 334
- ENV: 2 new files, now a total of 91
- GSS: 1 new file, now a total of 300
- PAT: 5 new files, now a total of 251
- PRI: 1 less file, now a total of 53
- TSA: 34 new files, now a total of 228
- VRT: 1 less file, now a total of 60
For downloading purposes, please keep in mind that the uncompressed GenBank flat files require approximately 771 GB (sequence files only); the ASN.1 data require approximately 633 GB.
More information about GenBank release 213.0, including current and upcoming changes like the change from GI sequence identifiers to accession.version, is available in the release notes.
- Epigenomics database to be retired June 1, 2016
- NCBI launches new Twitter account for NCBI Bookshelf
- New NCBI Insights blog post: Fast Sequence Inspection with ORFfinder and SmartBLAST (PubMed Labs)
- RefSeq release 76 is now available
- Genome Browsers section added to Gene
- Sequence Viewer version 3.14 upgrades platform
- New NCBI Variation summary page highlights all organisms in dbSNP or dbVar with full assembly annotations
- NCBI launches web-based iCn3D, a new viewer for 3D macromolecular structures
- NCBI annotates 300th organism with the Eukaryotic Genome Annotation Pipeline
- Genome Workbench 2.10.5 now available
- Preview the new BLAST home page!
- NCBI and RCSB PDB to assist ISCB in Sequence-Structure hackathon at ISMB Orlando 2016
- May 18th webinar: Using VDB BLAST Clients to Search Whole Genome Shotgun Contigs (WGS) and Transcriptome Shotgun Assembly (TSA) Data at the NCBI
- New NCBI video on YouTube: ProSplign comes to Genome Workbench
- New NCBI video on YouTube: Submitting BioSample Data to NCBI
- GenBank release 213.0 is now available via FTP
- NCBI News, May 2016 - NCBI NewsNCBI News, May 2016 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
