NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: Improvements to NCBI Services Promote Discovery
In an effort to make the full potential of the NCBI Web services and underlying databases more available to users, the NCBI has begun a long-term project to improve the relevance and usefulness of search results. This effort is called the Discovery Initiative. A primary goal of the Discovery Initiative is to promote the discovery of previously hidden relationships in the large amount of pre-calculated similarity data and pre-compiled links between different molecular and literature databases available at the NCBI. Changes in search interfaces and result displays are being phased in gradually and include the appearance of database ads, alternative search suggestions, and various sensors that will bring to the surface results in other databases that may be more relevant to the search. Particular Discovery components will appear in a context-specific manner ultimately producing the most relevant result possible. To help with this effort NCBI is also designing Web interfaces and links so that the effectiveness and popularity of these changes can be measured and studied. In turn, the results of these studies will be used to improve NCBI’s services even more.
Discovery Components in PubMed
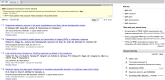
Many Discovery-related changes can already be seen in the PubMed database, and more will be coming soon. Current Discovery components appearing on PubMed results pages may include Related Queries, Title Searches, a Review tab, the Gene Sensor, and Recent Activity as shown in Figure 1. The Abstract Plus page view may include the top five Related Articles with review articles highlighted, Patient Drug Information and Recent Activity (Figure 2).
The Related Queries component shown under “Also try …” in the search results is a completely new kind of feature offering suggested queries from the most popular PubMed queries that contain the current search term. Using these suggested queries may provide more precise results than the current search.
Certain Discovery components improve and make more obvious previously existing pathways that are powerful but may have been cryptic before. For instance, the results available for ‘Title Searches’ and the ‘Review’ tab have been available by field-limited searches (term[Title], term AND review[Publication Type]). But, despite its usefulness, only a small minority of PubMed searchers use fielded searching.
Likewise, exposing the top five related articles the Abstract Plus view provides a more obvious alternative to the Related Articles available in the Links menu. Related Articles has been removed from the summary view of the search results, but is still available in the Abstract Plus record view. The new feature that highlights review articles here is a popular enhancement called ‘Recent Activity’ that provides a gateway to the broader literature relevant to a particular field. The Recent Activity component partially replaces the functions of the History tab allowing navigation to previous searches and record views in PubMed. Unlike the History tab however, Recent Activity provides access to searches and record views in other Entrez databases.
Other Discovery components such as the Gene Sensor, triggered when a gene symbol is used in a search, show results from other databases that may be more directly relevant than those from the current database. In some cases these sensors may provide useful results when the current database does not. For example, searching with the human gene symbol STK40 produces no results in PubMed itself. The Gene Sensor, however, reports results for the search of STK40 in the Gene database and provides a link to the human, mouse, rat, and the complete search results in Gene. These gene records have links to a variety of molecular data including mRNA and genomic sequences, genomic regions and maps, expression information, homologs in other species, and in the case of the human gene, a literature citation when PubMed found none.
Upcoming Changes
Many new Discovery-related changes will be appearing over the next several months. Discovery components will be released initially to a fraction of users so that NCBI can measure the effect and popularity of changes to the system. Some changes will appear first in PubMed and will be implemented in other databases as appropriate. The Recent Activity component, for example, was recently ported to some of the molecular databases after a test period in PubMed. In some cases, components will be database-specific such as the Taxonomy report that appears in the right-hand column of Entrez sequence database search results. In all cases the goal of these changes is to improve the usability and quality of results obtained from NCBI services.
Summary
The NCBI Website is the premier portal to biomedical literature and molecular biology data. Interconnecting the literature and these data is an enormously rich set of similarity and linkage relationships where previously unknown connections are waiting to be uncovered. While access to these connections has always been possible, it’s clear that many visitors are not enjoying the full potential of the system. Recent and upcoming changes to the NCBI Web experience will help expose these connections and promote discovery of the most biologically significant records and relationships in the NCBI databases.
New Databases and Tools
Bookshelf
The Bookshelf has added three new books: BLAST: Command Line Applications User Manual, The Intolerable Burden of Malaria: A New Look at the Numbers, and The Intolerable Burden of Malaria III: Progress and Perspectives. Books can be found at: www.ncbi.nlm.nih.gov/sites/entrez?db=Books.
Genome Build
Build 1.1 of Physomitrella patens (moss) is available in the Genomes database and on the NCBI Map Viewer. The Map Viewer page for this organism is: www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=3218
Microbial Genomes
Twenty-six finished microbial genomes were released between December 19 and January 14. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 169.0 is available via web and FTP that includes information as of December 11, 2008. A new release will be available in February 2009. A new linetype, DBLINK, will be implemented in GenBank files beginning with the February 2009 release. More information can be found in Section 1.4.1 of the GenBank Release Notes. Release notes can be found on the ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
Updates and Enhancements
RefSeq
RefSeq Release 33 is now available. This full release incorporates genomic, transcript, and protein data available as of January 16, 2009 and includes 10,325,282 records from 7,773 different organisms. The RefSeq website is: http://www.ncbi.nlm.nih.gov/RefSeq/. RefSeq data are also available through FTP: ftp.ncbi.nih.gov/refseq/release/.
Short Read Archive
The Short Read Archive, or SRA, is now available for searching in the Entrez system. “SRA” has been added as a choice in the Entrez search pulldown menu.
PubMed
PubMed has added a Recent Activity feature to the PubMed Abstract display page. This feature displays recent searches performed and the number of results found for those searches. The Recent Activity box also appears in some other Entrez database results pages with the databases being displayed within the box. This feature can be turned off if unwanted.
The PubMed and MeSH databases have been updated with the 2009 MeSH vocabulary.
HomoloGene
HomoloGene now offers multiple sequence alignments (MSAs) generated using MUSCLE. MSAs of genes and their homologs can be viewed by choosing a HomoloGene cluster, and either clicking on the “Show Multiple Alignment” link, or by choosing “Multiple Alignment” from the display menu. The HomoloGene Web resource is: www.ncbi.nlm.nih.gov/sites/entrez?db=homologene.
BLAST
Binaries for BLAST 2.2.19 are available from the ftp site: ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/. Some changes include: the BLASTDB environment variable now supports multiple database search paths; a smaller protein table is available to improve performance; and ‘formatrpsdb’ supports creation of databases larger than two gigabytes.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are described on the Announcement List summary page. Announce lists provide email announcements about changes and updates to NCBI resources. www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, February 2009 - NCBI NewsNCBI News, February 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...