NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: Primer-BLAST—NCBI’s Primer Designer and Specificity Checker
NCBI now offers Primer-BLAST, a Web service for designing target specific oligonucleotide primers for use in polymerase chain reaction (PCR) protocols. Primer-BLAST may be accessed from the NCBI BLAST Homepage or through the direct URL:
http://www.ncbi.nlm.nih.gov/tools/primer-blast/
This service integrates primer designing code from the popular Primer3 1 software with a specificity check that uses a custom BLAST search. Primer-BLAST eliminates the need to design primers at another site and then perform a search with those primers using the main NCBI BLAST service to check specificity. Moreover, Primer-BLAST can design primers that amplify only a particular splice variant of a gene – an important feature for PCR protocols measuring tissue specific expression. Primer-BLAST has a number of additional enhancements that make it better at picking specific primers than Primer3 and NCBI BLAST used separately.
Primer-BLAST Input
The Primer-BLAST interface contains elements of both the Primer3 tool and the NCBI BLAST Web service. The submission form is divided into three sections with commonly adjusted settings relevant to the target template, the primers, and the background specificity check. As with other BLAST services, the more specialized options are available under an expandable section at the bottom of the submission form, called “Advanced parameters” in Primer-BLAST.
Template
The target template sequence or accession number to be analyzed is placed in the large text area in the PCR Template section of the form. Specific binding regions on the template may be specified using the range boxes. With just the target template, Primer-BLAST will design primer pairs that are specific to the target template and unique in the target database.
Primers
One or both primers may be supplied in the Primer Parameters section. The most important and commonly modified primer-specific parameters such as desired product size, Tm ranges and Tm difference may be modified here as well. Primers may accompany a template, but may also be used without a template. With only a primer pair, Primer-BLAST identifies potential templates in the target database. In this case, the results are more precise than a specificity check with ordinary BLAST and have additional PCR condition parameters such as melting temperature (Tm) provided in the results.
Specificity
Parameters that may be adjusted in the Specificity Checking section of the form affect the nature of the background database and the level of stringency for avoiding nonspecific matches. The database specificity check does not simply find BLAST matches but includes adjustable mismatch cutoffs that focus on the 3′ end of the binding site to more effectively avoid mispriming. The most informative results are obtained by selecting the target database and organism that best match the template sequence source. The organism selection is managed by typing the name of the organism of interest in the “Organism” box. An auto-complete feature will find the closest match in the NCBI taxonomy database.
Four BLAST nucleotide databases are available for searching: RefSeq mRNA, Genome (selected reference assemblies), Genome (all chromosomes), and nr (the standard non-redundant database). The first two databases will give the most precise results as these are the best annotated and analyzed sets of sequences. In particular, if an NCBI mRNA Reference Sequence is used as a template with the RefSeq mRNA database, Primer-BLAST will design primers specific to the single splice variant represented by the reference sequence. The well characterized genome sequences of human, chimpanzee, mouse, rat, cow, dog, chicken, zebrafish, fruit fly, honeybee, Arabidopsis, and rice are available as the selected reference assemblies. These selected reference assemblies will produce results that are less redundant and easier to interpret when using genomic templates from these organisms. The remaining Genome database (all chromosomes) contains all chromosome Reference Sequences (NC_ accessions) including the selected reference assemblies, as well as their alternative assemblies, and chromosome records for other organisms, most notably microbial genome sequences. Finally, the nr database is available for the widest coverage of organisms.
Advanced Parameters
The Advanced Parameters section contains numerous adjustable settings affecting primer binding and efficacy from the Primer3 program. Two additional features that are only available in Primer-BLAST are options to avoid making primers that match repetitive or regions with biased base composition (low complexity regions) and an option to avoid regions that contain reported nucleotide polymorphisms (SNPs) from NCBI’s SNP database. As with Primer3, Primer-BLAST also has an option to design an internal hybridization probe for each product that can be used in various product detection assays.
Example
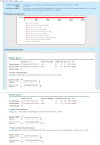
Using Primer-BLAST to design primers to distinguish the two transcripts of the human uracil-DNA glycosylase genes (UNG, GeneID: 7374) demonstrates the utility and precision of Primer BLAST (Figure 1). The UNG products code for enzymes involved in removing misincorpated uracil during DNA replication. The longer UNG1 transcript (NM_003362 ) codes for a mitochondrial enzyme while the shorter UNG2 transcript (NM_080911) that lacks the portion coding for the mitochondrial targeting sequence apparently functions in the nuclear genome of replicating cells. The top panel of Figure 1 shows the Primer-BLAST results that are specific to the UNG2 splice variant. In this case the NCBI reference sequence was used as a template with RefSeq mRNA database limited to human. By default the service designs only UNG2 specific primers with the reference sequence as the template. These conditions may be relaxed by checking the option for “Allow primer to amplify mRNA splice variants.” The bottom panel of Figure 1 shows that under these conditions new primers are found that will amplify both transcripts from the UNG gene. Expanding the database coverage provides a means for checking primers in additional species. The results using the nr database predict that many of the primer pairs designed against the human UNG transcripts should also amplify transcripts from other primate species (not shown).
Summary
Primer-BLAST provides a new more powerful and more comprehensive primer design tool by combining oligonucleotide primer design with specific genome and transcriptome level searches, and will save time and money by eliminating some of the trial and error of PCR primer design.
Reference
- 1. Steve Rozen and Helen J. Skaletsky (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365-386 [PubMed: 10547847]
New Databases and Tools
BLAST 2 Sequences
The NCBI BLAST web pages have a new option to align a query against a set of target sequences, rather than a BLAST database. This option allows you to align your query to one or more subject sequences and still use the standard BLAST web interface to optimize your search and change algorithm parameters. Each search is assigned a "Request ID" (RID) and is also listed under the "Recent Results" tab that you can access from the BLAST home page. The results are formatted as a standard BLAST report, except a "Dot Matrix view" (a "dot-plot" like graphic of the alignments) is available in the new report design if only one subject sequence was searched. Links to BLAST web pages can be found at: blast.ncbi.nlm.nih.gov/BLAST.cgi Step-by-step instructions can be found at: blast.ncbi.nlm.nih.gov/docs/align_seqs.pdf
My NCBI
My NCBI version 2.0 was released in September with new features and updated functions. New features include: account and username options; navigation, preferences, and filter improvements; My Saved Searches tool; and My Collections tool. Another new feature, My Bibliography, allows authors to search and collect citations for their publications. For a full description of new and improved features, please see the NLM Technical Bulletin article at: www.nlm.nih.gov/pubs/techbull/so08/so08_myncbi_redesign.html . My NCBI is located at: www.ncbi.nlm.nih.gov/sites/myncbi/
BarSTool
The Barcode of Life project is one in which DNA barcode sequences are being collected from vouchered specimens of all biological species. The idea is to make it easier to identify biological specimens via a short gene sequence from a standardized position in the genome. NCBI has created a submission tool, Barcode Submission Tool (BarSTool), to submit barcode sequences to GenBank. For more information about BarSTool or the Barcode of Life project see: www.ncbi.nlm.nih.gov/Genbank/barcode.html.
Bookshelf
The Bookshelf has added two new books entitled: Hepatitis C Viruses: Genomes and Molecular Biology and NCBI News. Books can be found at: www.ncbi.nlm.nih.gov/sites/entrez?db=Books.
Microbial Genomes
Thirty-two finished microbial genomes were released (August 15-October 10). The original sequence data files submitted to GenBank/EMBL/DDBJ can be found at: ftp://ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are available via FTP at: ftp://ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 167.0 is available via web and FTP that includes information as of August 19, 2008.
Updates and Enhancements
Discovery PubMed Pages
PubMed Summary results pages have begun showing related information from other resources on the right side of the page. Drug Sensor, an NCBI tool that detects drug names in a search, was released in August. As of September, additional resources will be shown to a percentage of users to aid in the discovery process.
RefSeq
RefSeq Release 31 is available via web and FTP. This full release incorporates genomic, transcript, and protein data available as of August 30, 2008 and includes 9,145,702 records, 5,859,648 proteins, and sequences from 5,513 different organisms.
The RefSeq website is: www.ncbi.nlm.nih.gov/RefSeq/.
Genome Assembly
Genome annotation for Ciona intestinalis build 1.1 and Arabidopsis thaliana build 8.1 were released in September. For more annotation updates and links to Genome Resource pages see: www.ncbi.nlm.nih.gov/Genomes/.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are described on the Announcement List summary page. Announce lists provide email announcements about changes and updates to NCBI resources. http://www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. http://www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- PubMedLinks to PubMed
- NCBI News, November 2008 - NCBI NewsNCBI News, November 2008 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...