NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-.
This publication is provided for historical reference only and the information may be out of date.
Summary
The resources provided by NCBI for studying the three-dimensional (3D) structures of proteins center around two databases: the Molecular Modeling Database (MMDB), which provides structural information about individual proteins; and the Conserved Domain Database (CDD), which provides a directory of sequence and structure alignments representing conserved functional domains within proteins(CDs). Together, these two databases allow scientists to retrieve and view structures, find structurally similar proteins to a protein of interest, and identify conserved functional sites.
To enable scientists to accomplish these tasks, NCBI has integrated MMDB and CDD into the Entrez retrieval system (Chapter 15). In addition, structures can be found by BLAST, because sequences derived from MMDB structures have been included in the BLAST databases (Chapter 16). Once a protein structure has been identified, the domains within the protein, as well as domain “neighbors” (i.e., those with similar structure) can be found. For novel data not yet included in Entrez, there are separate search services available.
Protein structures can be visualized using Cn3D, an interactive 3D graphic modeling tool. Details of the structure, such as ligand-binding sites, can be scrutinized and highlighted. Cn3D can also display multiple sequence alignments based on sequence and/or structural similarity among related sequences, 3D domains, or members of a CDD family. Cn3D images and alignments can be manipulated easily and exported to other applications for presentation or further analysis.
Overview
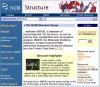
The Structure homepage (Figure 1) contains links to the more specialized pages for each of the main tools and databases, introduced below, as well as search facilities for the Molecular Modeling Database (MMDB; Ref. 1).
MMDB is based on the structures within Protein Data Bank (PDB) and can be queried using the Entrez search engine, as well as via the more direct but less flexible Structure Summary search (see Figure 1). Once found, any structure of interest can be viewed using Cn3D (2), a piece of software that can be freely downloaded for Mac, PC, and UNIX platforms.
Often used in conjunction with Cn3D is the Vector Alignment Search Tool (VAST; Refs. 3, 4). VAST is used to precompute “structure neighbors” or structures similar to each MMDB entry. For people that have a set of 3D coordinates for a protein not yet in MMDB, there is also a VAST search service. The output of the precomputed VAST searches is a list of structure records, each representing one of the Non-Redundant PDB chain sets (nr-PDB), which can also be downloaded. There are four clustered subsets of MMDB that compose nr-PDB, each consisting of clusters having a preset level of sequence similarity.
The structures within MMDB are now being linked to the NCBI Taxonomy database (Chapter 4). Known as the PDBeast project, this effort makes it possible to find: (1) all MMDB structures from a particular organism; and (2) all structures within a node of the taxonomy tree (such as lizards or Bacillus), which launches the Taxonomy Browser showing the number of MMDB records in each node.
The second database within the Structure resources is the Conserved Domain Database (CDD; Ref. 5), originally based largely on Pfam and SMART, collections of alignments that represent functional domains conserved across evolution. CDD now also contains the alignments of the NCBI COG database, the NCBI Library of Ancient Domains (LOAD) along with new curated alignments assembled at NCBI. CDD can be searched from the CDD page in several ways, including by a domain keyword search. Three tools have been developed to assist in analysis of CDD: (1) the CD-Search, which uses a BLAST-based algorithm to search the position-specific scoring matrices (PSSM) of CDD alignments; (2) the CD-Browser, which provides a graphic display of domains of interest, along with the sequence alignment; and (3) the Conserved Domain Architecture Retrieval Tool (CDART), which searches for proteins with similar domain architectures.
All the above databases and tools are discussed in more detail in other parts of this Chapter, including tips on how to make the best use of them.
Content of the Molecular Modeling Database (MMDB)
Sources of Primary Data
To build MMDB (1), 3D structure data are retrieved from the PDB database (6) administered by the Research Collaboratory for Structural Bioinformatics (RCSB). In all cases, the structures in MMDB have been determined by experimental methods, primarily X-ray crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy. Theoretical structure models are omitted. The data in each record are then checked for agreement between the atomic coordinates and the primary sequence, and the sequence data are then extracted from the coordinate set. The resulting agreement between sequence and structure allows the record to be linked efficiently into searches and alignment displays involving other NCBI databases.
The data are converted into ASN.1 (7), which can be parsed easily and can also accept numerous annotations to the structure data. In contrast to a PDB record, a MMDB record in ASN.1 contains all necessary bonding information in addition to sequence information, allowing consistent display of the 3D structure using Cn3D. The annotations provided in the PDB record by the submitting authors are added, along with uniformly defined secondary structure and domain features. These features support structure-based similarity searches using VAST. Finally, two coordinate subsets are added to the record: one containing only backbone atoms, and one representing a single-conformer model in cases where multiple conformations or structures were present in the PDB record. Both of these additions further simplify viewing both an individual structure and its alignments with structure neighbors in Cn3D. When this process is complete, the record is assigned a unique Accession number, the MMDB-ID (Box 1), while also retaining the original four-character PDB code.
Box 1
Accession numbers.
Annotation of 3D Domains
After initial processing, 3D domains are automatically identified within each MMDB record. 3D domains are annotations on individual MMDB structures that define the boundaries of compact substructures contained within them. In this way, they are similar to secondary structure annotations that define the boundaries of helical or β-strand substructures. Because proteins are often similar at the level of domains, VAST compares each 3D domain to every other one and to complete polypeptide chains. The results are stored in Entrez as a Related 3D Domain link.
To identify 3D domains within a polypeptide chain, MMDB's domain parser searches for one or more breakpoints in the structure. These breakpoints fall between major secondary structure elements such that the ratio of intra- to interdomain contacts remains above a set threshold. The 3D domains identified in this way provide a means to both increase the sensitivity of structure neighbor calculations and also present 3D superpositions based on compact domains as well as on complete polypeptide chains. They are not intended to represent domains identified by comparative sequence and structure analysis, nor do they represent modules that recur in related proteins, although there is often good agreement between domain boundaries identified by these methods.
Links to Other NCBI Resources
After initially processing the PDB record, structure staff add a number of links and other information that further integrate the MMDB record with other NCBI resources. To begin, the sequence information extracted from the PDB record is entered into the Entrez Protein and/or Nucleotide databases as appropriate, providing a means to retrieve the structure information from sequence searches. As with all sequences in Entrez, precomputed BLAST searches are then performed on these sequences, linking them to other molecules of similar sequence. For proteins, these BLAST neighbors may be different than those determined by VAST; whereas VAST uses a conservative significance threshold, the structural similarities it detects often represent remote relationships not detectable by sequence comparison. The literature citations in the PDB record are linked to PubMed so that Entrez searches can allow access to the original descriptions of the structure determinations. Finally, semiautomatic processing of the “source” field of the PDB record provides links to the NCBI Taxonomy database. Although these links normally follow the genus and species information given, in some cases this information is either absent in the PDB record or refers only to how a sample was obtained. In these cases, the staff manually enters the appropriate taxonomy links.
The MMDB Record
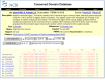
The Structure Summary page for each MMDB record summarizes the database content for that record and serves as a starting point for analyzing the record using the NCBI structure tools (Figure 2).
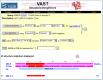
VAST Structure Neighbors
Although VAST itself is not a database, the VAST results computed for each MMDB record are stored with this record and are summarized on a separate page for the whole polypeptide chain as well as for each 3D domain found in the protein (Figure 3). These pages can be accessed most easily by clicking on either the chain bar or the 3D Domain bar in the graphic display of the Structure Summary page (Figure 2).
nr-PDB
The non-redundant PDB database (nr-PDB) is a collection of four sets of sequence-dissimilar cluster PDB polypeptide chains assembled by NCBI Structure staff. The four sets differ only in their respective levels of non-redundancy. The staff assembles each set by comparing all the chains available from PDB with each other using the BLAST algorithm. The chains are then clustered into groups of similar sequence using a single-linkage clustering procedure. Chains within a sequence-similar group are automatically ranked according to the quality of their structural data. Details of the measures used to determine structure precision and completeness and the methodology of assembling the nr-PDB clusters can be found on the nr-PDB Web page.
Content of the Conserved Domain Database (CDD)
What Is a Conserved Domain (CD)?
CDs are recurring units in polypeptide chains (sequence and structure motifs), the extents of which can be determined by comparative analysis. Molecular evolution uses such domains as building blocks and these may be recombined in different arrangements to make different proteins with different functions. The CDD contains sequence alignments that define the features that are conserved within each domain family. Therefore, the CDD serves as a classification resource that groups proteins based on the presence of these predefined domains. CDD entries often name the domain family and describe the role of conserved residues in binding or catalysis. Conserved domains are displayed in MMDB Structure summaries and link to a sequence alignment showing other proteins in which the domain is conserved, which may provide clues on protein function.
Sources of Primary Data
The collections of domain alignments in the CDD are imported either from two databases outside of the NCBI, named Pfam (8) and SMART (9); from the NCBI COB database; from another NCBI collection named LOAD; and from a database curated by the CDD staff. The first task is to identify the underlying sequences in each collection and then link these sequences to the corresponding ones in Entrez. If the CDD staff cannot find the Accession numbers for the sequences in the records from the source databases, they locate appropriate sequences using BLAST. Particular attention is paid to any resulting match that is linked to a structure record in MMDB, and the staff substitute alignment rows with such sequences whenever possible. After the staff imports a collection, they then choose a sequence that best represents the family. Whenever possible, the staff chooses a representative that has a structure record in MMDB.
The Position-specific Score Matrix (PSSM)
Once imported and constructed, each domain alignment in CDD is used to calculate a model sequence, called a consensus sequence, for each CD. The consensus sequence lists the most frequently found residue in each position in the alignment; however, for a sequence position to be included in the consensus sequence, it must be present in at least 50% of the aligned sequences. Aligned columns covered by the consensus sequence are then used to calculate a PSSM, which memorizes the degree to which particular residues are conserved at each position in the sequence. Once calculated, the PSSM is stored with the alignment and becomes part of the CDD. The RPS-BLAST tool locates CDs within a query sequence by searching against this database of PSSMs.
Reverse Position-specific BLAST (RPS-BLAST)
RPS-BLAST (Chapter 16) is a variant of the popular Position-specific Iterated BLAST (PSI-BLAST) program. PSI-BLAST finds sequences similar to the query and uses the resulting alignments to build a PSSM for the query. With this PSSM the database is scanned again to draw in more hits and further refine the scoring model. RPS-BLAST uses a query sequence to search a database of precalculated PSSMs and report significant hits in a single pass. The role of the PSSM has changed from “query” to “subject”; hence, the term “reverse” in RPS-BLAST. RPS-BLAST is the search tool used in the CD-Search service.
The CD Summary
Analogous to the Structure Summary page, the CD Summary page displays the available information about a given CD and offers various links for either viewing the CD alignment or initiating further searches (Figure 4). The CD Summary page can be retrieved by selecting the CD name on any page.
CD Records Curated at NCBI
In 2002, NCBI released the first group of curated CD records, a new and expanding set of annotated protein multiple sequence alignments and corresponding structure alignments. These new records have Accession numbers beginning with “cd” and have been added to the default CD-Search database. Most curated CD records are based on existing family descriptions from SMART and Pfam, but the alignments may have been revised extensively by quantitatively using three-dimensional structures and by re-examining the domain extent. In addition, CDD curators annotate conserved functional residues, ligands, and co-factors contained within the structures. They also record evidence for these sites as pointers to relevant literature or to three-dimensional structures exemplifying their properties. These annotations may be viewed using Cn3D and thus provide a direct way of visualizing functional properties of a protein domain in the context of its three-dimensional structure. (See Box 3 and Figure 7.)
Box 3
Example query: finding and viewing CDs in a protein.
The Distinction between 3D Domains and CDs
The term “domain” refers in general to a distinct functional and/or structural unit of a protein. Each polypeptide chain in MMDB is analyzed for the presence of two classes of domains, and it is important for users to understand the difference between them. One class, called 3D Domains, is based solely on similar, compact substructures, whereas the second class, called Conserved Domains (CDs), is based solely on conserved sequence motifs. These two classifications often agree, because the compact substructures within a protein often correspond to domains joined by recombination in the evolutionary history of a protein. Note that CD links can be identified even when no 3D structures within a family are known. Moreover, 3D Domain links may also indicate relationships either to structures not included in CDD entries or to structures so distantly related that no significant similarity can be found by sequence comparisons.
Finding and Viewing Structures
For an example query on finding and viewing structures, see Box 2.
Box 2
Example query: finding and viewing structural data of a protein.
Why Would I Want to Do This?
- To determine the overall shape and size of a protein
- To locate a residue of interest in the overall structure
- To locate residues in close proximity to a residue of interest
- To develop or test chemical hypotheses regarding an enzyme mechanism
- To locate or predict possible binding sites of a ligand
- To interpret mutation studies
- To find areas of positive or negative charge on the protein surface
- To locate particularly hydrophobic or hydrophilic regions of a protein
- To infer the 3D structure and related properties of a protein with unknown structure from the structure of a homologous protein
- To study evolutionary processes at the level of molecular structure
- To study the function of a protein
- To study the molecular basis of disease and design novel treatments
How to Begin
The first step to any structural analysis at NCBI is to find the structure records for the protein of interest or for proteins similar to it. One may search MMDB directly by entering search terms such as PDB code, protein name, author, or journal in the Entrez Structure Search box on the Structure homepage. Alternative points of entry are shown below.
By using the full array of Entrez search tools, the resulting list of MMDB records can be honed, ideally, to a workable list from which a record can be selected. Users should note that multiple records may exist for a given protein, reflecting different experimental techniques, conditions, and the presence or absence of various ligands or metal ions. Records may also contain different fragments of the full-length molecule. In addition, many structures of mutant proteins are also available. The PDB record for a given structure generally contains some description of the experimental conditions under which the structure was determined, and this file can be accessed by selecting the PDB code link at the top of the Structure Summary page.
Alternative Points of Entry
Structure Summary pages can also be found from the following NCBI databases and tools:
- Select the Structure links to the right of any Entrez record found; records with Structure links can also be located by choosing Structure links from the Display pull-down menu.
- Select the Related Sequences link to the right of an Entrez record to find proteins related by sequence similarity and then select Structure links in the Display pull-down menu.
- From the results of any protein BLAST search, click on a red 'S' linkout to view the sequence alignment with a structure record.
Viewing 3D Structures
3D Domains
The 3D domains of a protein are displayed on the Structure Summary page. It is useful to know how many 3D domains a protein contains and whether they are continuous in sequence when viewing the full 3D structure of the molecule.
Secondary Structure
Knowing the secondary structure of a protein can also be a useful prelude to viewing the 3D structure of the molecule. The secondary structure can be viewed easily by first selecting the Protein link to the left of the desired chain in the graphic display. Finding oneself in Entrez Protein, selecting Graphics in the Display pull-down menu presents secondary structure diagrams for the molecule.
Full Protein Structures
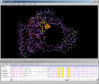
Cn3D is a software package for displaying 3D structures of proteins. Once it has been installed and the Internet browser has been configured correctly, simply selecting the View 3D Structure button on a Structure Summary page launches the application. Once the structure is loaded, a user can manipulate and annotate it using an array of options as described in the Cn3D Tutorial. By default, Cn3D colors the structure according to the secondary structure elements. However, another useful view is to color the protein by domain (see Style menu options), using the same color scheme as is shown in the graphic display on the Structure Summary page. These color changes also affect the residues displayed in the Sequence/Alignment Viewer, allowing the identification of domain or secondary structure elements in the primary sequence. In addition to Cn3D, users can also display 3D structures with RasMol or Mage. Structures can also be saved locally as an ASN.1, PDB, or Mage file (depending on the choice of structure viewer) for later display.
Finding and Viewing Structure Neighbors
For an example query on finding and viewing structure neighbors, see Box 2.
Why Would I Want to Do This?
- To determine structurally conserved regions in a protein family
- To locate the structural equivalent of a residue of interest in another related protein
- To gain insights into the allowable structural variability in a particular protein family
- To develop or test chemical hypotheses regarding an enzyme mechanism
- To predict possible binding sites of a ligand from the location of a binding site in a related protein
- To identify sites where conformational changes are concentrated
- To interpret mutation studies
- To find areas of conserved positive or negative charge on the protein surface
- To locate conserved hydrophobic or hydrophilic regions of a protein
- To identify evolutionary relationships across protein families
- To identify functionally equivalent proteins with little or no sequence conservation
How to Begin
The Vector Alignment Search Tool (VAST) is used to calculate similar structures on each protein contained in the MMDB. The graphic display on each Structure Summary page (Figure 2) links directly to the relevant VAST results for both whole proteins and 3D domains:
- Selecting the chain bar displays the VAST Structure Neighbors page for the entire chain.
Alternative Points of Entry
- From any Entrez search, select Related 3D Domains to the right of any record found to view the Vast Structure Neighbors page.
Viewing a 2D Alignment of Structure Neighbors
A graphic 2D HTML alignment of VAST neighbors can be viewed as follows:
- On the View/Save bar, configure the pull-down menus to the right of the View Alignment button.
- Select View Alignment.
Viewing a 3D Alignment of Structure Neighbors
Alignments of VAST structure neighbors can be viewed as a 3D image using Cn3D.
- On the View/Save bar, configure the pull-down menus to the right of the View 3D Structure button.
- Select View 3D Structure.
Cn3D automatically launches and displays the aligned structures. Each displayed chain has a unique color; however, the portions of the structures involved in the alignment are shown in red. These same colors are also reflected in the Sequence/Alignment Viewer. Among the many viewing options provided by Cn3D, of particular use is the Show/Hide menu that allows only the aligned residues to be viewed, only the aligned domains, or all residues of each chain.
Finding and Viewing Conserved Domains
For an example query on finding and viewing conserved domains, see Box 3.
Why Would I Want to Do This?
- To locate functional domains within a protein
- To predict the function of a protein whose function is unknown
- To establish evolutionary relationships across protein families
- To interpret mutation studies
- To predict the structure of a protein of unknown structure
How to Begin
Following the Domains link for any protein in Entrez, one can find the conserved domains within that protein. The CD-Search (or Protein BLAST, with CD-Search option selected) can be used to find conserved domains (CDs) within a protein. Either the Accession number, gi number, or the FASTA sequence can be used as a query.
Alternative Points of Entry
Information on the CDs contained within a protein can also be found from these databases and tools:
- From any Entrez search: select the Domains link to the right of a displayed record.
- From an Entrez Domains search: choose Domains from the Entrez Search pull-down menu and enter a search term to retrieve a list of CDs. Clicking on any resulting CD displays the CD Summary page. To find the location of this CD in an aligned protein, select the CD link following a protein name in the bottom portion of this page.
Viewing Conserved Domains
Results from a CD search are displayed as colored bars underneath a sequence ruler. Moving the mouse over these bars reveals the identity of each domain; domains are also listed in a format similar to BLAST summary output (Chapter 16). Pairwise alignments between the matched region of the target protein and the representative sequence of each domain are shown below the bar. Red letters indicate residues identical to those in the representative sequence, whereas blue letters indicate residues with a positive BLOSUM62 score in the BLAST alignment.
Viewing Multiple Alignments of a Query Protein with Members of a Conserved Domain
These can be displayed by clicking a CD bar within a MMDB Structure Summary page or from a hyperlinked CD name on a CD-Search results page.
Viewing CD Alignments in the Context of 3D Structure
If members of a CD have MMDB records, one of these records can be viewed as a 3D image along with the sequence alignment using Cn3D (launched by selecting the pink dot on a CD-Search results page). As in other alignment views, colored capital letters indicate aligned residues, allowing the sequence of the protein sequence of interest to be mapped onto the available 3D structure.
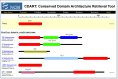
Finding and Viewing Proteins with Similar Domain Architectures
For an example query on finding and viewing proteins with similar domain architectures, see Box 3.
Why Would I Want to Do This?
- To locate related functional domains in other protein families
- To gain insights into how a given CD is situated within a protein relative to other CDs
- To explore functional links between different CDs
- To predict the function of a protein whose function is unknown
- To establish evolutionary relationships across protein families
How to Begin
Following the Domain Relatives link for any protein in Entrez, one can find other proteins with similar domain architecture. The Conserved Domain Architecture Retrieval Tool (CDART) can take an Accession number or the FASTA sequence as a query to find out the domain architecture of a protein sequence and list other proteins with related domain architectures.
Alternative Points of Entry
Results of a CDART Search
These are described in Figure 5. The protein “hits”, which have similar domain architectures to the query sequence, can be further refined by taxonomic group, in which the results can be limited to selected nodes of the taxonomic tree. Furthermore, search results may be limited to those that contain only particular conserved domains.
Links Between Structure and Other Resources
Integration with Other NCBI Resources
As illustrated in the sections above, there are numerous connections between the Structure resources and other databases and tools available at the NCBI. What follows is a listing of major tools that support connections.
Entrez
Because Entrez is an integrated database system (Chapter 15), the links attached to each structure give immediate access to PubMed, Protein, Nucleotide, 3D Domain, CDD, or Taxonomy records.
BLAST
Although the BLAST service is designed to find matches based solely on sequence, the sequences of Structure records are included in the BLAST databases, and by selecting the PDB search database, BLAST searches only the protein sequences provided by MMDB records. A new Related Structure link provides 3D views for sequences with structure data identified in a BLAST search.
BLink
The BLink report represents a precomputed list of similar proteins for many proteins (see, for example, links from Entrez Gene records; Chapter 19). The 3D Structures option on any BLink report shows the BLAST hits that have 3D structure data in MMDB, whereas the CDD-Search button displays the CD-Search results page for the query protein.
Microbial Genomes
A particularly useful interface with the structural databases is provided on the Microbial Genomes page (10). To the left of the list of genomes are several hyperlinks, two of which offer users direct access to structural information. The red [D] link displays a listing of every protein in the genome, each with a link to a BLink page showing the results of a BLAST pdb search for that protein. The [S] link displays a similar protein list for the selected genome, but now with a listing of the conserved domains found in each protein by a CD-Search.
Links to Non-NCBI Resources
The Protein Data Bank (PDB)
As stated elsewhere, all records in the MMDB are obtained originally from the Protein Data Bank (PDB) (6). Links to the original PDB records are located on the Structure Summary page of each MMDB record. Updates of the MMDB with new PDB records occur once a month.
Pfam and SMART
The CDD staff imports CD collections from both the Pfam and SMART databases. Links to the original records in these databases are located on the appropriate CD Summary page. Both Pfam and SMART are updated several times per year in roughly bimonthly intervals, and the CDD staff update CDD accordingly.
Saving Output from Database Searches
Exporting Graphics Files from Cn3D
Structures displayed in Cn3D can be exported as a Portable Network Graphics (PNG) file from within Cn3D (the Export PNG command in the File menu). The structure file itself, in the orientation currently being viewed, can also be saved for later launching in Cn3D.
FTP
MMDB
Users can download the NCBI Structure databases from the NCBI FTP site: ftp://ftp.ncbi.nih.gov/mmdb. A Readme file contains descriptions of the contents and information about recent updates. Within the mmdb directory are four subdirectories that contain the following data:
CDD
CDD data can be downloaded from ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd. A Readme file contains descriptions of the data archives. Users can download the PSSMs for each CD record, the sequence alignments in mFASTA format, or a text file containing the accessions and descriptions of all CD records.
Frequently Asked Questions
References
- 1.
- Wang Y , Anderson JB , Chen J , Geer LY , He S , Hurwitz DI , Liebert CA , Madej T , Marchler GH , Marchler-Bauer A , Panchenko AR , Shoemaker BA , Song JS , Thiessen PA , Yamashita RA , Bryant SH . MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2002;30:249–252. [PMC free article: PMC99072] [PubMed: 11752307]
- 2.
- Wang Y , Geer LY , Chappey C , Kans JA , Bryant SH . Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. [PubMed: 10838572]
- 3.
- Madej T , Gibrat J-F , Bryant SH . Threading a database of protein cores. Proteins. 1995;23:356–369. [PubMed: 8710828]
- 4.
- Gibrat J-F , Madej T , Bryant SH . Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. [PubMed: 8804824]
- 5.
- Marchler-Bauer A , Panchenko AR , Shoemaker BA , Thiessen PA , Geer LY , Bryant SH . CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. [PMC free article: PMC99109] [PubMed: 11752315]
- 6.
- Westbrook J , Feng Z , Jain S , Bhat TN , Thanki N , Ravichandran V , Gilliland GL , Bluhm W , Weissig H , Greer DS , Bourne PE , Berman HM . The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. [PMC free article: PMC99110] [PubMed: 11752306]
- 7.
- Ohkawa H , Ostell J , Bryant S . MMDB: an ASN.1 specification for macromolecular structure. Proc Int Conf Intell Syst Mol Biol. 1995;3:259–267. [PubMed: 7584445]
- 8.
- Bateman A , Birney E , Cerruti L , Durbin R , Etwiller L , Eddy SR , Griffiths-Jones S , Howe KL , Marshall M , Sonnhammer ELL . The Pfam proteins family database. Nucleic Acids Res. 2002;30:276–280. [PMC free article: PMC99071] [PubMed: 11752314]
- 9.
- Letunic I , Goodstadt L , Dickens NJ , Doerks T , Schultz J , Mott R , Ciccarelli F , Copley RR , Ponting CP , Bork P . SMART: a Web-based tool for the study of genetically mobile domains. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. [PMC free article: PMC99073] [PubMed: 11752305]
- 10.
- Wang Y , Bryant S , Tatusov R , Tatusova T . Links from genome proteins to known 3D structures. Genome Res. 2000;10:1643–1647. [PMC free article: PMC310938] [PubMed: 11042161]
- Summary
- Overview
- Content of the Molecular Modeling Database (MMDB)
- Content of the Conserved Domain Database (CDD)
- Finding and Viewing Structures
- Finding and Viewing Structure Neighbors
- Finding and Viewing Conserved Domains
- Finding and Viewing Proteins with Similar Domain Architectures
- Links Between Structure and Other Resources
- Saving Output from Database Searches
- FTP
- Frequently Asked Questions
- References
- Macromolecular Structure Databases - The NCBI HandbookMacromolecular Structure Databases - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...