NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: Re-designed PopSet
NCBI's PopSet database of related sequences and alignments from phylogenetic, population, mutation, and ecosystem studies has been completely redesigned and now features an embedded graphical alignment and better integration of related data from other PopSets and other Entrez databases. The new pages also include on-the-fly analysis with BLAST and Tree View.
The PopSet Record View
The PopSet record view is now fully integrated with the updated Entrez system and can be addressed simply with the PopSet database name and the identifier as shown below.
http://www.ncbi.nlm.nih.gov/popset/298351991
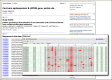
The record display shown in Figure 1 consists of up to three sections: the study details showing the article reporting the current set; a list of the sequence records in the set; and, when available, the submitted alignment displayed in the embedded Graphical Sequence Viewer (GSV), now also appearing in Entrez Gene and SNP record views. The PopSet embedded alignment view shows the alignment portion of the full GSV display of the master or top sequence in the multiple-alignment. Clicking on the “Open full-view” link opens the GSV nucleotide view of the top sequence showing the detailed alignment tracks.
As in the other Entrez databases, the “Display Setting” menu controls the format of the records displayed; the “Send to” menu manages saving data, shown in Figure 2. Display options are similar to those available for the Nucleotide database and include the standard sequence formats such as FASTA and GenBank. The sequence record formats are presented within the PopSet display rather than by linking to the sequence database.
The “Send to” menu can send data to the Entrez clipboard, Collections in a My NCBI account, or to a file on the local computer. The file saving format options include the standard sequence formats, popular multiple alignment formats – FASTA plus gap, CLUSTAL, Nexus, and Phylip – are also available making the alignments easy to use for local analysis.
Improved PopSet-PopSet Connections
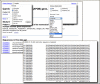
PopSet now features more explicit connections between PopSets associated with the same study. As always, following the link from a PubMed record retrieves all PopSets for molecules used in the study. In the previous version of PopSet, however, it was not easy to navigate from one PopSet to others that are part of the same study. The PopSet-PopSet link now provides rapid access to related PopSets. The related PopSets also are listed “Other data sets from this study” in the right-hand Discovery Column of the full record. Figure 3 shows the items in the Discovery Column and the corresponding related data in PopSet and PubMed.
Analysis Tools: BLAST and Tree View
For PopSets with fewer than 100 sequences, analysis tools are available at the top of the right- hand Discovery Column (Figure 3). These allow generating or re-generating an alignment with BLAST or, if a submitted alignment is present, displaying a distance tree (Tree View) based on the alignment. Figure 4 shows the results of the BLAST and Tree View tools for a phylogenetic study set that has a submitted alignment. The link to run BLAST is especially useful in cases where the set does not contain a submitted alignment, for example PopSet: 338197537. In such cases the Tree View can be invoked after running the BLAST alignment through the “Distance tree of results” link on the BLAST output.
Summary
The NCBI PopSet database has been fully updated to the new Entrez system and includes new record displays and better access to related information. These improvements will make the growing collection of PopSets easier to access, download, and analyze.
New My NCBI Interface
My NCBI now has customizable modules making it even easier to manage your NCBI preferences, collections, bibliographies, saved searches, and more. A video highlighting the new homepage and features is on the NCBI YouTube Channel.
Transcriptome Shotgun Assembly (TSA) Database Available for BLAST
The Transcriptome Shotgun Assembly (TSA) BLAST database is now available from the database list for the main NCBI BLAST services. TSA is an archive of computationally assembled mRNA sequences from primary data such as Expressed Sequence Tag (EST) and raw sequence reads. These sequences were previously a part of the BLAST nucleotide nr (nt) database but have been moved because of their increasing numbers and special characteristics. The TSA page has more information on the nature and sources of TSA sequences.
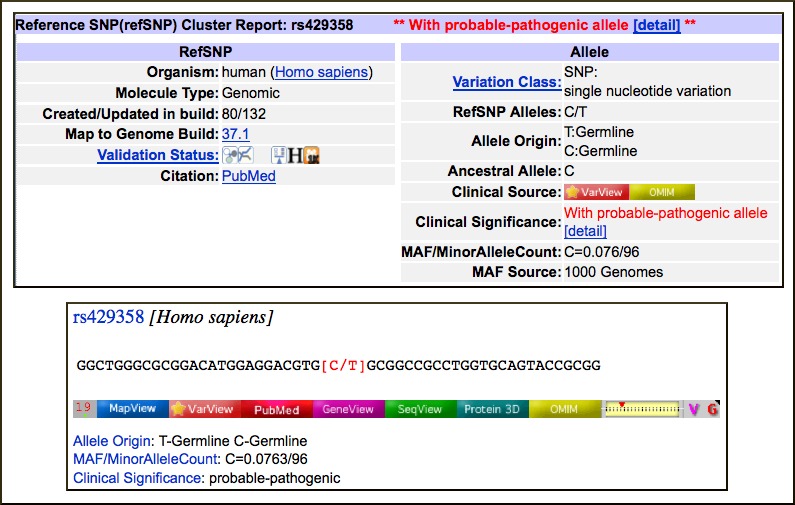
New Attributes for Human Variants in dbSNP
New attributes related to allele origin, clinical significance, and population genetics are available in dbSNP. These attributes allow searching and filtering of human variations for the characteristics listed below.
- Allele Origin: Summarizes the reported origin(s) of the variant allele asserted by each submitter for the submitted SNP (ss). Current values are germline, somatic, and unknown. Additional attributes will be added in the future including not-tested, tested-inconclusive, and other.
- Clinical significance: Reports potential health impact of the allele. Possible values:
- unknown
- untested
- non-pathogenic
- probable-non-pathogenic
- probable-pathogenic
- pathogenic
- drug response
- histocompatibility
- other
- Global minor allele frequency (MAF): Shows the minor allele frequency for each RefSNP included in a default global population. Since this is being provided to distinguish common polymorphism from rare variants, the MAF is actually the second most frequent allele value. For example, if there are 3 alleles with frequencies of 0.50, 0.49, and 0.01, the MAF will be reported as 0.49. The current default global population is 1000Genome phase 1 genotype data from 629 worldwide individuals, released in the 08-04-2010 dataset.
- Suspect: Variation suspected to be false positive due to various artifacts.These new attributes are shown in the images below for the rs429358 Cluster Report and Document Summary.
Please see the online help for more information and more examples.
Updated BLAST Genome Search Pages
The genome-specific BLAST pages linked to the top of the NCBI BLAST homepage and accessible from the Map Viewer homepage now use the standard BLAST form with genome specific databases. This change eliminates the older separate interface and provides the full functionality of the standard BLAST interface including the ability to adjust all algorithm parameters, the capability to edit and re-submit searches, to sort descriptions and alignments in the output, and the full range of formatting and downloading options.
NLM Contest: Show off your Apps! Invitation to Submit Applications that Work with NLM Biomedical Data
The National Library of Medicine (NLM) is challenging people to create innovative software applications that use the Library's vast collection of biomedical data. The purpose of this contest is to foster the development of innovative software applications that will further NLM’s mission of aiding the dissemination and exchange of scientific and other information pertinent to medicine and public health. Winners will be recognized at an awards ceremony at the National Library of Medicine and links to their application will be publicized on NLM Web sites. The NLM "Show Off Your Apps" Challenge is open to individuals over the age of 18, teams of individuals, and organizations in the United States. Eligible software applications must make use of NLM’s vast collection of biomedical data including downloadable data sets, application programming interfaces, and/or software tools. The challenge.gov website has detailed information on the contest.
Applications should be submitted to the challenge.gov site by August 31, 2011.
New Videos on NCBI’s YouTube Channel
In addition to the video introducing the new My NCBI mentioned above, four other instructional videos recently became available on NCBI’s YouTube channel:
The Sequence Read and Trace Archive Databases to Continue
Recently, NCBI announced that the Sequence Read Archive (SRA) and Trace Archive repositories would be discontinued due to budget constraints (NCBI News, March 2011). However, with the commitment of interim funding and a plan for future support developed in collaboration with other NIH Institutes and NIH grantees, NCBI will now continue to accept submissions and maintain the Sequence Read Archive (SRA) and Trace Archive repositories for high-throughput sequence data. These repositories will now focus on high-throughput data that support other kinds of data at the NCBI including:
- RNA-Seq, ChIP-Seq, and epigenomic data that are submitted to GEO
- Genomic and Transcriptomic assemblies that are submitted to GenBank
- 16S ribosomal RNA data associated with metagenomics that are submitted to GenBank
The full announcement on the NCBI site has more details.
BLAST 2.2.25+ Release and New Set-up Instructions
Stand-alone BLAST+ (v2.2.25) is now on the FTP site. Improvements include hard-masking of databases, faster formatting of databases using makeblastdb, XML and best hit options for Blast2Sequences, multiple query psiblast, selection of any master sequence in psiblast with multiple alignment input, and query and subject length in tabular output. The BLAST News has more detailed information on changes. Detailed set-up instructions for standalone BLAST are now a part of the BLAST User Manual on the NCBI Bookshelf.
Microbial Genomes Update
One hundred thirty five finished microbial genomes were released between March 1 and May 31, 2011. The original sequence data files submitted to GenBank/EMBL/DDBJ are available in the Bacteria directory in the /genbank/genomes area of the GenBank FTP site. One hundred twelve RefSeq provisional versions were made from a selected set of finished genomes. These are available from the /genomes/Bacteria directory on the FTP site.
In addition, 305 microbial whole genome shotgun-sequencing projects were added to GenBank during this period. The original submitted files are available in the Bacteria_DRAFT directory in the GenBank genomes area. RefSeq provisional versions of 84 of these projects are available in the /genomes/Bacteria_DRAFT area of the FTP site.
All GenBank and RefSeq microbial genomes are incorporated in the NCBI integrated Entrez search and retrieval system and the BLAST sequence similarity search service.
RefSeq News
RefSeq Release 47 is available through Entrez, BLAST, and the RefSeq FTP site. The current release includes 17.6 million sequence records from 12,000 organisms. Release notes provide more detailed information.
GenBank News
GenBank release 183 is available through the NCBI web and FTP sites. The current release incorporates data available as of Apr 11, 2011 and, with the whole-genome shotgun portion, contains 317,952,894,329 bases from 198,156,212 sequence records. Release notes describe the current state of data and upcoming changes.
NCBI Discovery Workshops at Washington University: July 26-27, 2011
NCBI will present a two-day workshop on July 26 and 27th, at Washington University in St. Louis, Missouri. The course is free and is open to anyone interested in NCBI resources. The workshops provide hands-on experience exploring practical examples using tools and databases on the NCBI website. The four workshops are Sequences, Genomes, and Maps; Proteins, Domains and Structures; NCBI BLAST Services; and Human Variation and Disease Genes. The Discovery Workshops page has more information.
Announce Lists and RSS Feeds
Eighteen topic-specific mailing lists are available that provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page. Subscribe to the NCBI Announce list to receive updates on the NCBI News.
Twelve RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce.
NCBI’s Facebook page and Twitter feed also provide updates on NCBI resources.
Send comments and questions about NCBI resources to info@ncbi.nlm.nih.gov, or call 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- Featured Resource: Re-designed PopSet
- New My NCBI Interface
- Transcriptome Shotgun Assembly (TSA) Database Available for BLAST
- New Attributes for Human Variants in dbSNP
- Updated BLAST Genome Search Pages
- NLM Contest: Show off your Apps! Invitation to Submit Applications that Work with NLM Biomedical Data
- New Videos on NCBI’s YouTube Channel
- The Sequence Read and Trace Archive Databases to Continue
- BLAST 2.2.25+ Release and New Set-up Instructions
- Microbial Genomes Update
- RefSeq News
- GenBank News
- NCBI Discovery Workshops at Washington University: July 26-27, 2011
- Announce Lists and RSS Feeds
- NCBI News, June 2011 - NCBI NewsNCBI News, June 2011 - NCBI News
- pokergosu-apt eggc.vipPH3 (40)BioProject
Your browsing activity is empty.
Activity recording is turned off.
See more...