NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: An Expanded Set of Discovery Components in the Entrez System
Several new features of the NCBI Entrez Web service are aspects of the ongoing Discovery Initiative described in the February and March 2009 Issues of the NCBI News. These new discovery components in the literature and sequence databases make the most relevant and interesting results more obvious and readily accessible.
There are three main categories of discovery components that now appear: Sensors, Database Ads, and Analysis Tools.
A sensor detects certain types of search terms and provides access to potentially more relevant results. For PubMed, new sensors include a Citation Sensor that is activated when someone searches with a literature citation and an Accession Sensor that provides a direct link to the sequence databases when someone searches with an NCBI sequence identifier. A variable type of sensor, the Hot Topic Sensor, also appears in PubMed. This new sensor that was inspired by the rapidly changing state of data for H1N1 influenza virus during the current outbreak appears for searches relevant to the recently added H1N1 viral sequences but in the future will be tailored to respond to other topical issues. The new more precise Gene Sensor that debuted in the PubMed database in January is now available in the protein and nucleotide databases.
A Database Ad promotes related information in other databases that may be more useful or may provide unexpected connections. New Database Ads in PubMed highlight the full-text PubMed Central database. The PubMed Central Ad that appears with PubMed results displays articles that are also available in full-text in the PubMed Central database. In the Abstract Plus View, the ads link to articles in PubMed Central that cite the PubMed record. A new Structure Ad appears in both the PubMed and sequence databases for articles that report a 3-D structure or for sequences derived from structure records. Viral Genome Resources Ads for influenza, dengue, SARS, and retroviruses such as HIV now appear in the sequence databases on sequence records of viral origin.
Analysis Tools that provide on-the-fly analysis are important components of the discovery initiative. Sequence analysis tools available for sequence records now include a direct link that will perform a BLAST search with the sequence as well as a link to run a conserved domain search for protein records. These new links accompany the direct link to design primers that has already been present on nucleotide records for several months.
All of these new discovery components are designed to help researchers find the most relevant information in the NCBI databases in the fewest mouse clicks.
Sensors in Entrez
As mentioned above, new sensors in Entrez include the Citation Sensor, the Accession Sensor, the Hot Topic Sensor for the H1N1 influenza virus, and the new Gene Sensor for sequence databases.
The new Citation Sensor automatically returns results from the PubMed Citation Matcher when it detects a query resembling a literature citation in a PubMed Search. Citation queries often retrieve irrelevant results when entered as a general PubMed search. The Citation Matcher service, now available as a part of the PubMed Advanced interface, is designed specifically for matching literature citations with PubMed records:
www.ncbi.nlm.nih.gov/pubmed/advanced
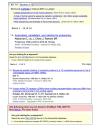
The Citation Sensor makes the power of the Citation Matcher more widely available. A minimal citation query would normally include an author name and a publication year or a journal name and publication year. For example, a search with “Lander 2001 Nature” quickly finds the Nature publication on the human genome sequence (Initial sequencing and analysis of the human genome sequence) as one of three articles found by the Citation Sensor (Figure 1, top panel). In comparison, the direct PubMed search retrieves 14 records, 11 of which are not from the journal Nature.
The Accession Sensor in PubMed is designed to provide relevant results when a PubMed search contains a sequence accession number. While GenBank sequence accession numbers reported in PubMed articles will find the source publication when used directly as a PubMed query, many accessions have no corresponding publication. Derivative sequence records such as NCBI Reference Sequences are often not associated directly with any PubMed records. Also, in many cases the goal of searching with accession identifiers is to find the sequence record itself and not the publication. In all of the above situations the accession sensor is quite useful in providing relevant results.
The middle panel of Figure 1 shows the results obtained in PubMed searching with a GenBank accession for the human dopamine D2 receptor (DRD2) mRNA (X51362). The search retrieves two PubMed citations that reference the accession as expected. The citation sensor in this case provides a convenient means to directly retrieve the sequence record without performing a separate search or following a link from one of the publications. The bottom panel of Figure 1 shows the results obtained using the corresponding NCBI Reference Sequence accession identifier for the DRD2 mRNA (NM_000795). There are no results found in PubMed since the RefSeq identifier is not cited in any publications or included in the abstract. However, the accession sensor provides access directly to the correct sequence record.
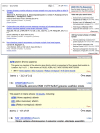
Another kind of sensor, the Hot Topic sensor, now appears in PubMed in response to increased searches related to the recent H1N1 influenza outbreak. In its present form, the sensor appears at the top of the right hand discovery column when it detects search terms that indicate interest in the H1N1 influenza sequences, and provides a link to the specialized H1N1 Influenza page described in the May, 2009 NCBI News (Figure 2, top panel). The Hot Topic Sensor will be deployed in different formats in response to current events in order to provide easy access to topical results.
The Gene Sensor that has been active in PubMed for several months is now in the protein and nucleotide databases. As in PubMed, the Gene Sensor is triggered by a gene symbol in a search. The older sequence database gene search feature remains active and will still return results from the gene database when the search does not trigger the Gene Sensor. The middle panel of Figure 2 shows the Gene Sensor triggered in the nucleotide database by a search with the mammalian gene symbol AFM. The sensor allows retrieval of relevant gene records with access to nucleotide and protein sequences while the direct nucleotide results contain large numbers of irrelevant matches. The gene search results triggered by a search with “afamin” shown in the bottom panel of Figure 2 also provide a better set of results than the direct nucleotide search.
Database Ads
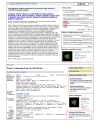
Two new Database Ads for the full-text PubMed Central database appear in PubMed. A link appears in all PubMed search results (Figure 2, top panel) displaying all articles that are also available in PubMed Central. Another ad for PubMed Central appears in the Abstract Plus record view and links to articles also in PubMed Central that cite the current article (Figure 3, top panel). This not only provides rapid access to full-text articles, but also offers another mechanism to expand the search to potentially related articles. As PubMed Central continues to expand the number of citations, it may also provide a useful measure of the significance of a particular article.
A Structure Ad now appears in both the PubMed and sequence database record views (Figure 3). This ad features a thumbnail image of 3-D molecular structures reported in the PubMed article or linked directly to the sequence record. The image is linked to the corresponding record in the structure database. From here the structure may be displayed and manipulated in NCBI’s Cn3D structure viewer. In the sequence databases, records for influenza, dengue viruses, SARS, and retroviruses like HIV now display an ad for the taxon-specific viral genome resources area of the NCBI Web site. An example of the ad is shown in the bottom panel of Figure 3 for an influenza virus sequence. The viral resources pages have collections of viral sequences, genotyping and other specialized tools that virus researchers may find more useful than those within the general Entrez.
Analysis Tools
Direct links to sequence analysis tools in sequence records provide a means to instantly generate sequence-specific reagents through Primer-BLAST and update the annotation on all nucleotide and protein records through the ability to perform a live BLAST or conserved domain database search (Figure 3, bottom panel). Up to 20% of NCBI BLAST searches use NCBI database identifiers or copy-pasted NCBI formatted sequences as queries; the direct link to BLAST now makes it much easier to perform BLAST searches with NCBI database records.
Summary
New Discovery components in the NCBI System – Sensors, Database Ads, and Analysis tools – make the Entrez system more powerful and easier to use by providing context sensitive results that traverse traditional database boundaries. These components not only make it possible to find relevant information in fewer steps but also help make more obvious unanticipated connections that are often essential to scientific discovery.
New Databases and Tools
BioSystems
NCBI BioSystems is a new database designed to aggregate biosystems information from collaborating public databases. BioSystems is a centralized repository of data that connects the biosystem records with associated literature, molecular, and chemical data throughout the Entrez system and facilitates computation on biosystems data. The NCBI BioSystems database currently contains biological pathways from the KEGG and BioCyc databases and is designed to accommodate other types of biosystems. Detailed diagrams and annotations for individual biosystems are available on the Web sites of the source databases. Links to Biosystems are now available from records in the NCBI Gene, HomoloGene, OMIM, and Protein Clusters databases. For more information, please see the BioSystems homepage: www.ncbi.nlm.nih.gov/biosystems/
Genome Resources
NCBI’s Genome Resource pages provide a comprehensive guide for a specific organism including links to NCBI resources as well as outside groups and consortia. New genome resource pages are available for the Pea Aphid (Acyrthosiphon pisum) and goat (Capra hircus). Links can be found under the “Organism-Specific” section of the Genomic Biology page: www.ncbi.nlm.nih.gov/Genomes/.
Microbial Genomes
Twenty-one finished microbial genomes were released between April 30 and May 28. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 171.0 is available via web and FTP. The current release includes information available as of April 10, 2009. Release notes are available on the on the NCBI ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
NCBI is considering ceasing support for index files. Affected GenBank users are encouraged to read that section of the release notes and provide feedback to the GenBank group.
Updates and Enhancements
RefSeq
RefSeq Release 35 is now available via Entrez and FTP. This full release incorporates genomic, transcript, and protein data available as of May 4, 2009. It includes 10,993,891 records from 8,393 different species and strains. Changes since the previous release can be found in the release notes on the FTP site. The RefSeq website is: www.ncbi.nlm.nih.gov/RefSeq/. The FTP site is: ftp.ncbi.nlm.nih.gov/refseq/release.
dbSNP
Complete data for the dbSNP Human build 130 are available on the FTP site and for searching on the web. More detailed genome build information is available on the dbSNP page: www.ncbi.nlm.nih.gov/SNP/snp_summary.cgi.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html. To receive updates on the NCBI News, please see: www.ncbi.nlm.nih.gov/About/news/announce_submit.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, June 2009 - NCBI NewsNCBI News, June 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...